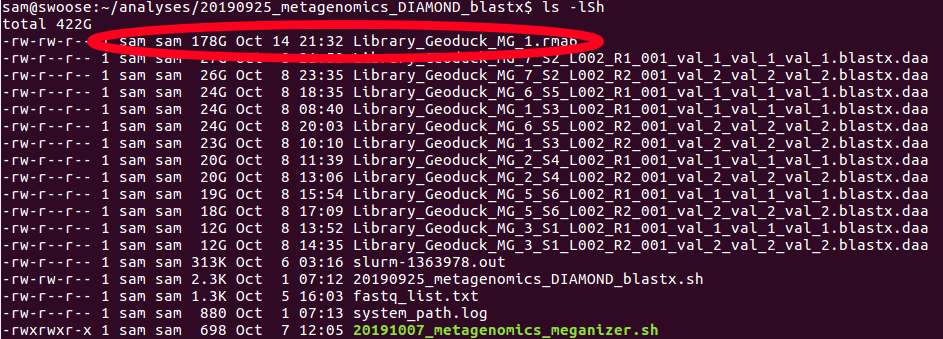
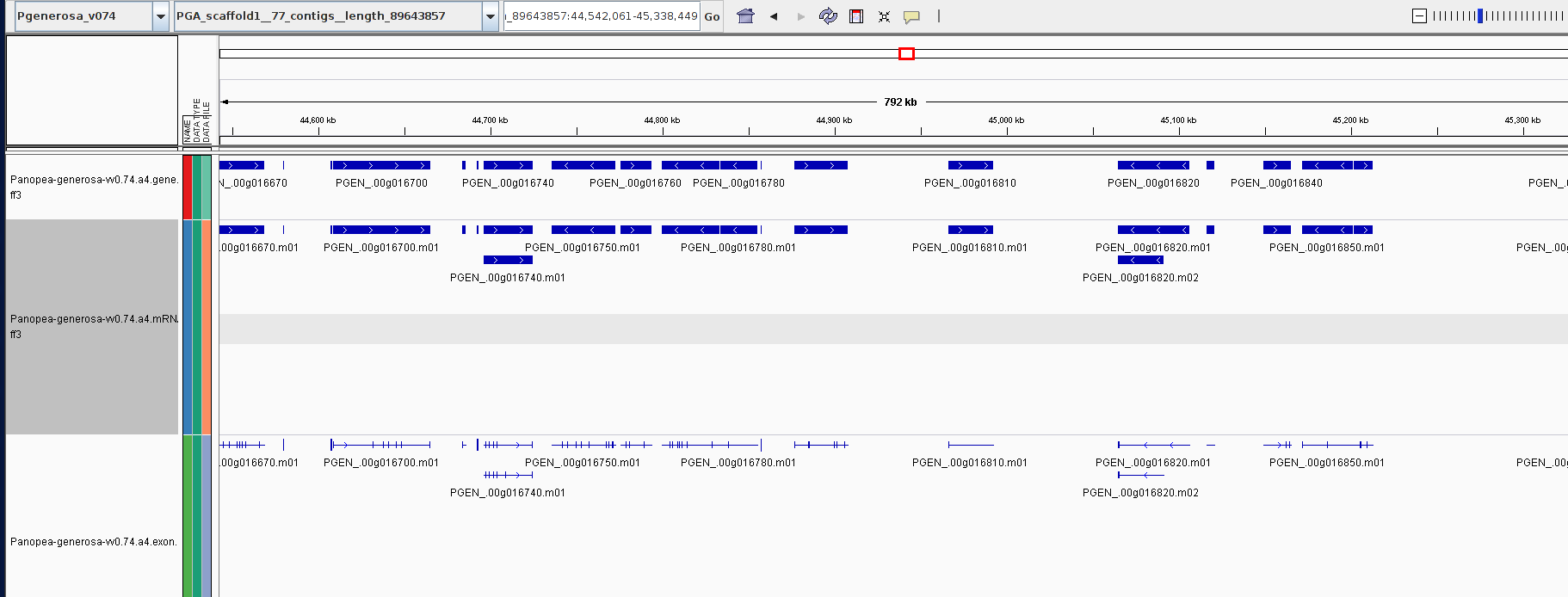
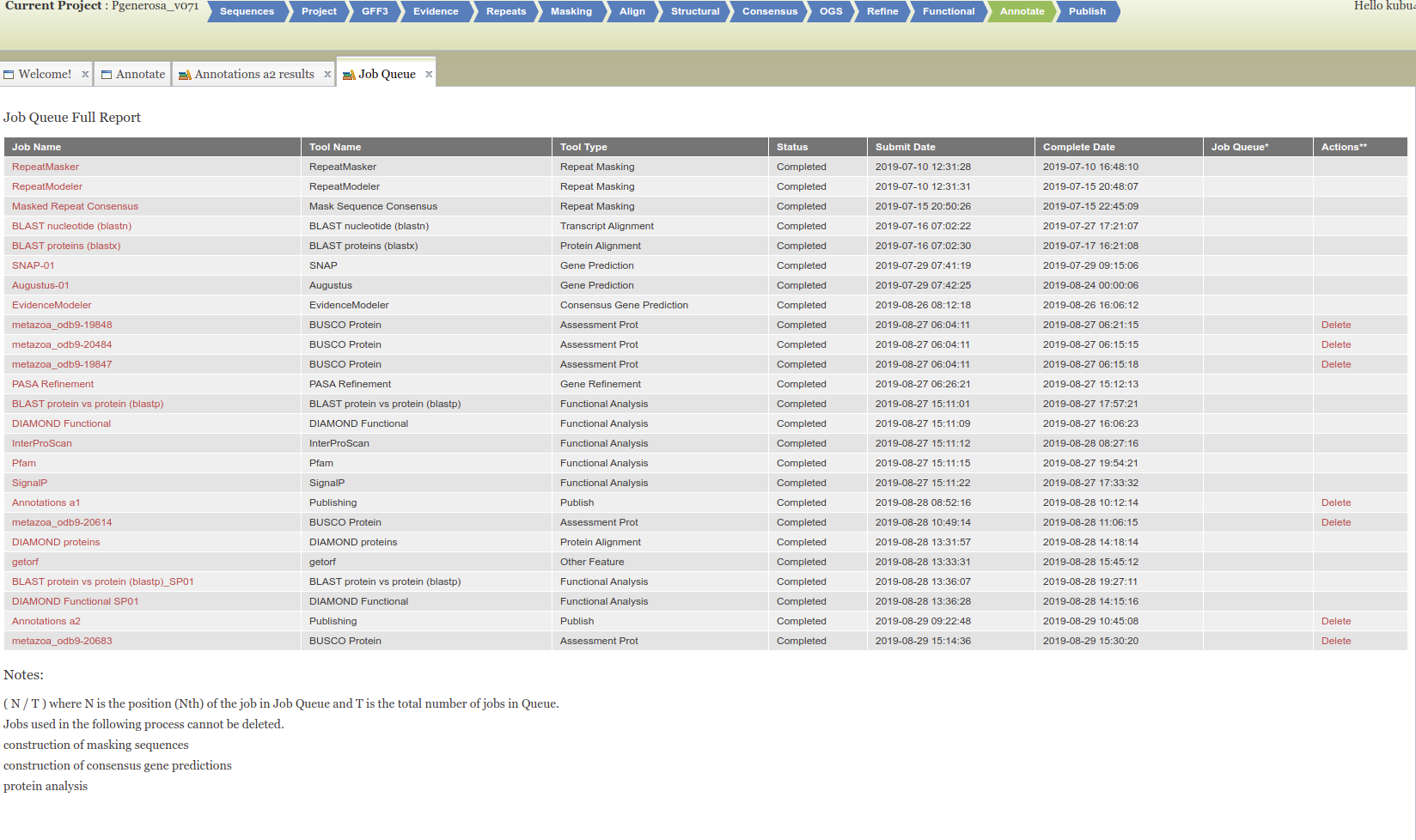
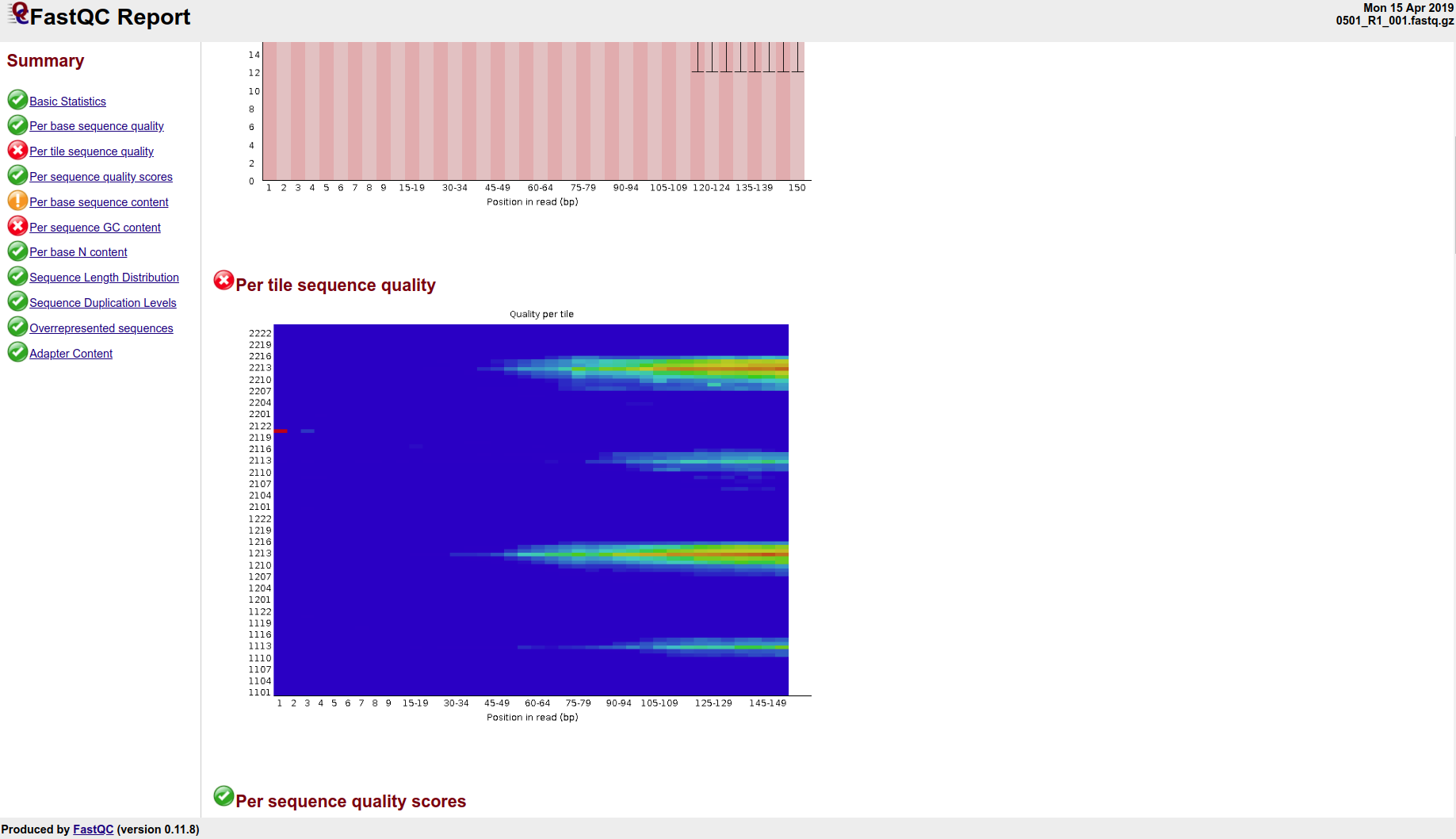
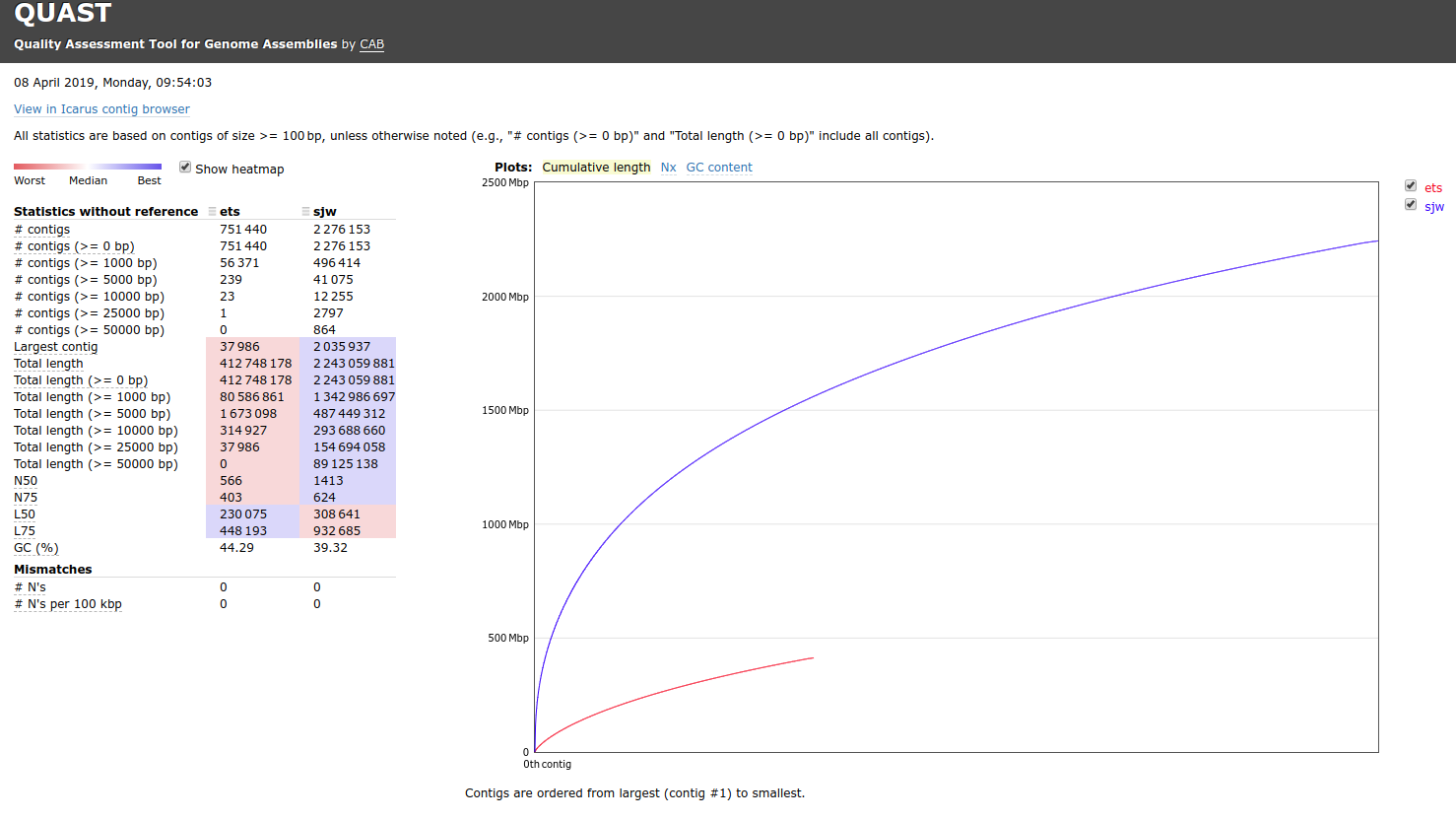
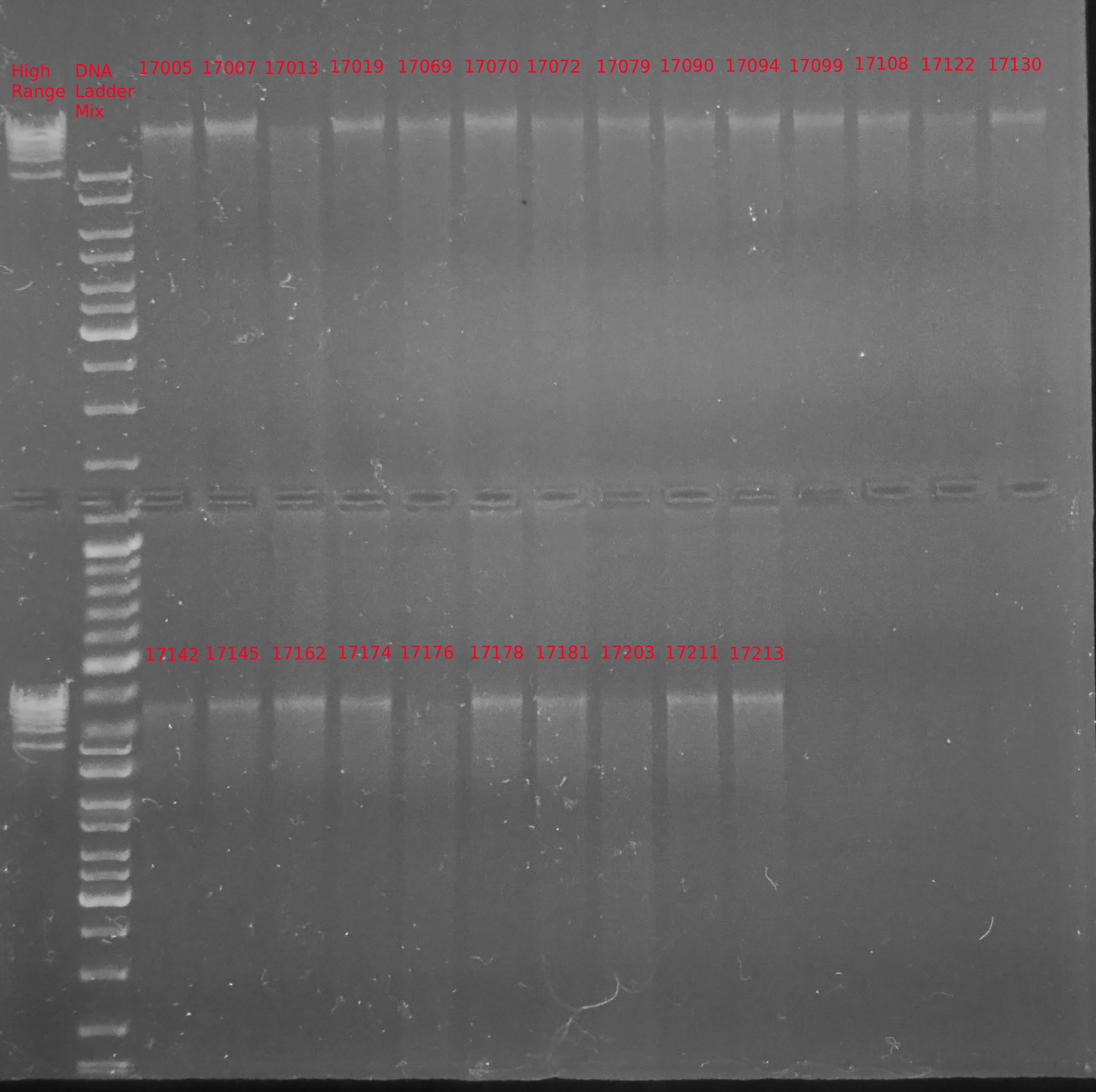
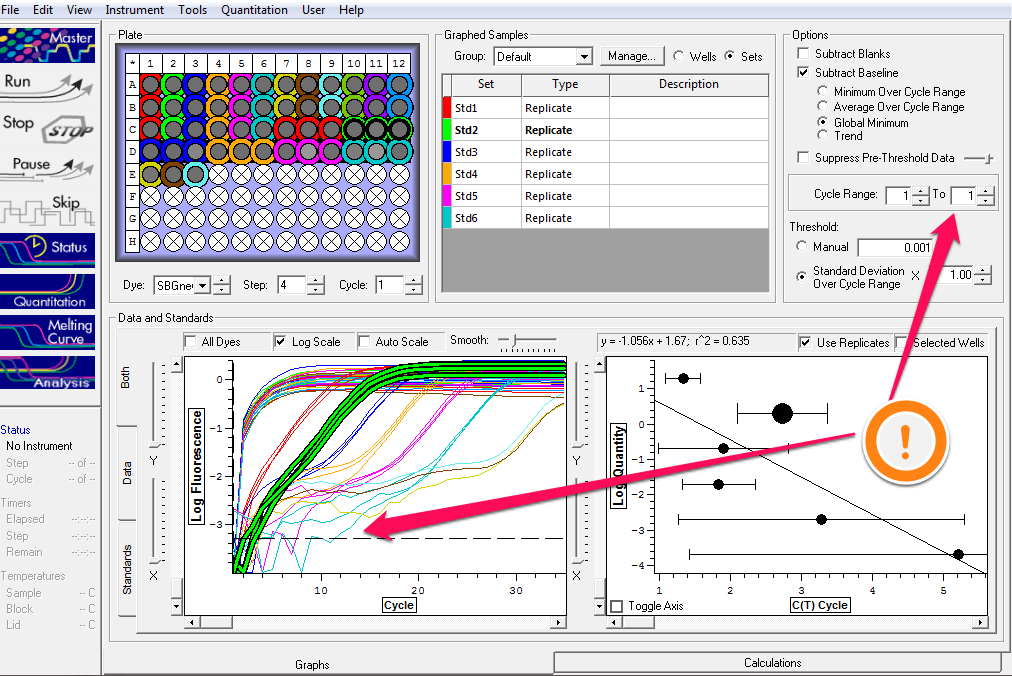
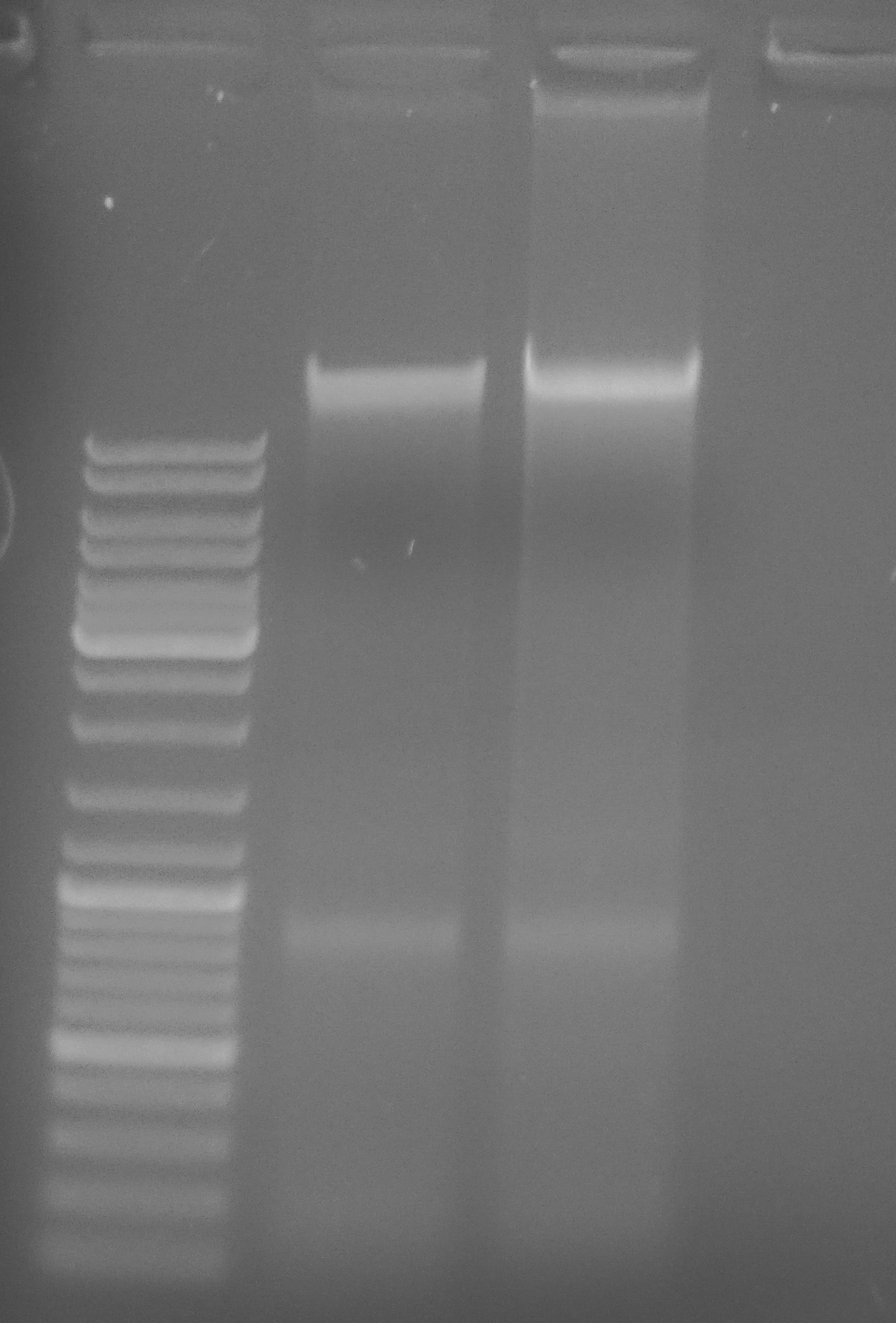
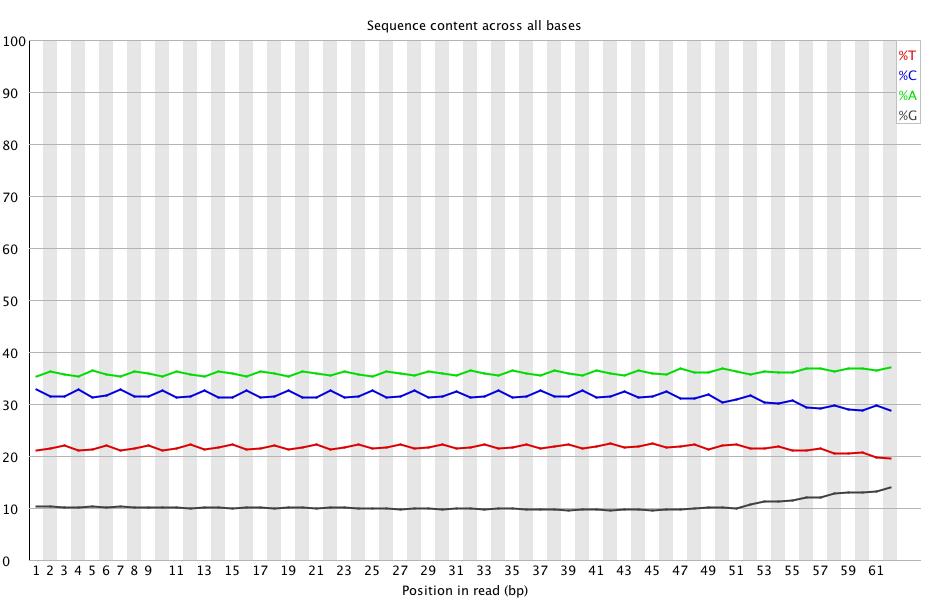
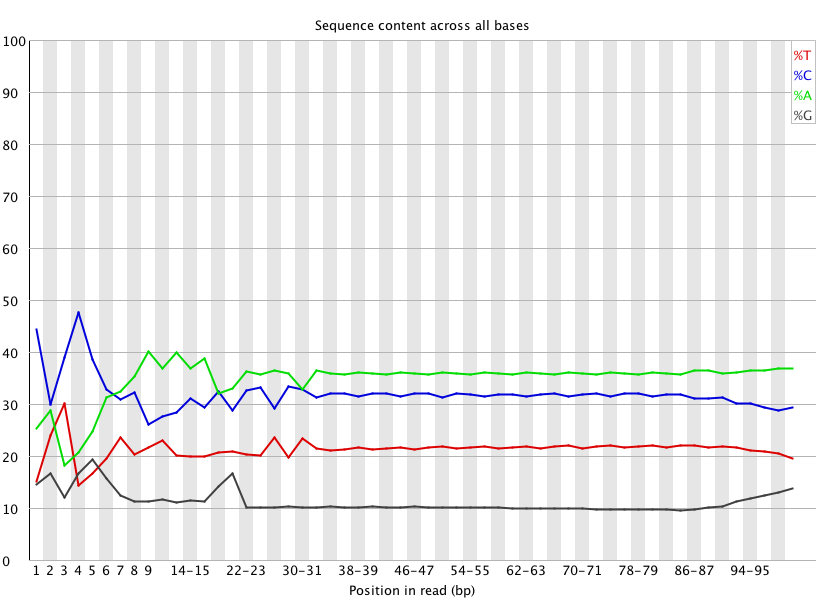
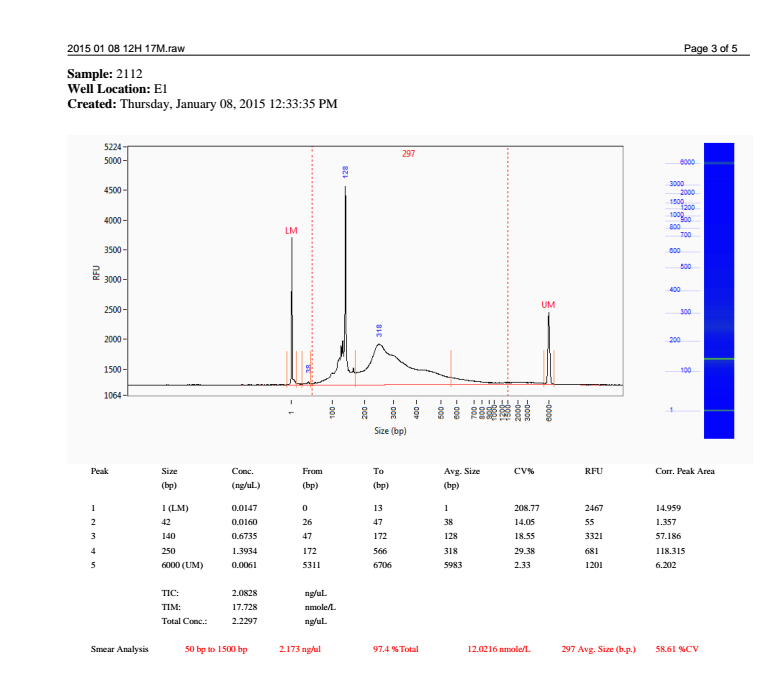
+
diff --git a/docs/index.xml b/docs/index.xml
index e1225d461..5408a72d5 100644
--- a/docs/index.xml
+++ b/docs/index.xml
@@ -10,7 +10,94 @@
quarto-1.3.450
-
Tue, 02 Jan 2024 08:00:00 GMT
+
Wed, 03 Jan 2024 08:00:00 GMT
+
-
+
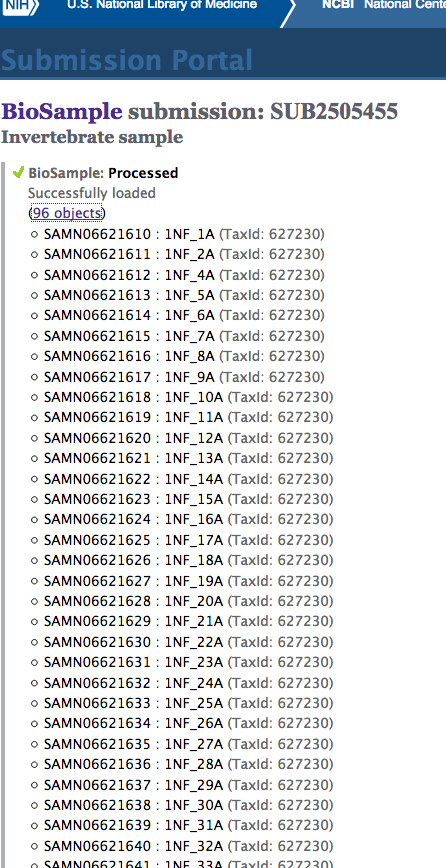
Data Exploration - CEABIGR Spurious Transcription Calculations and Plotting
+ Sam White
+
+Intro
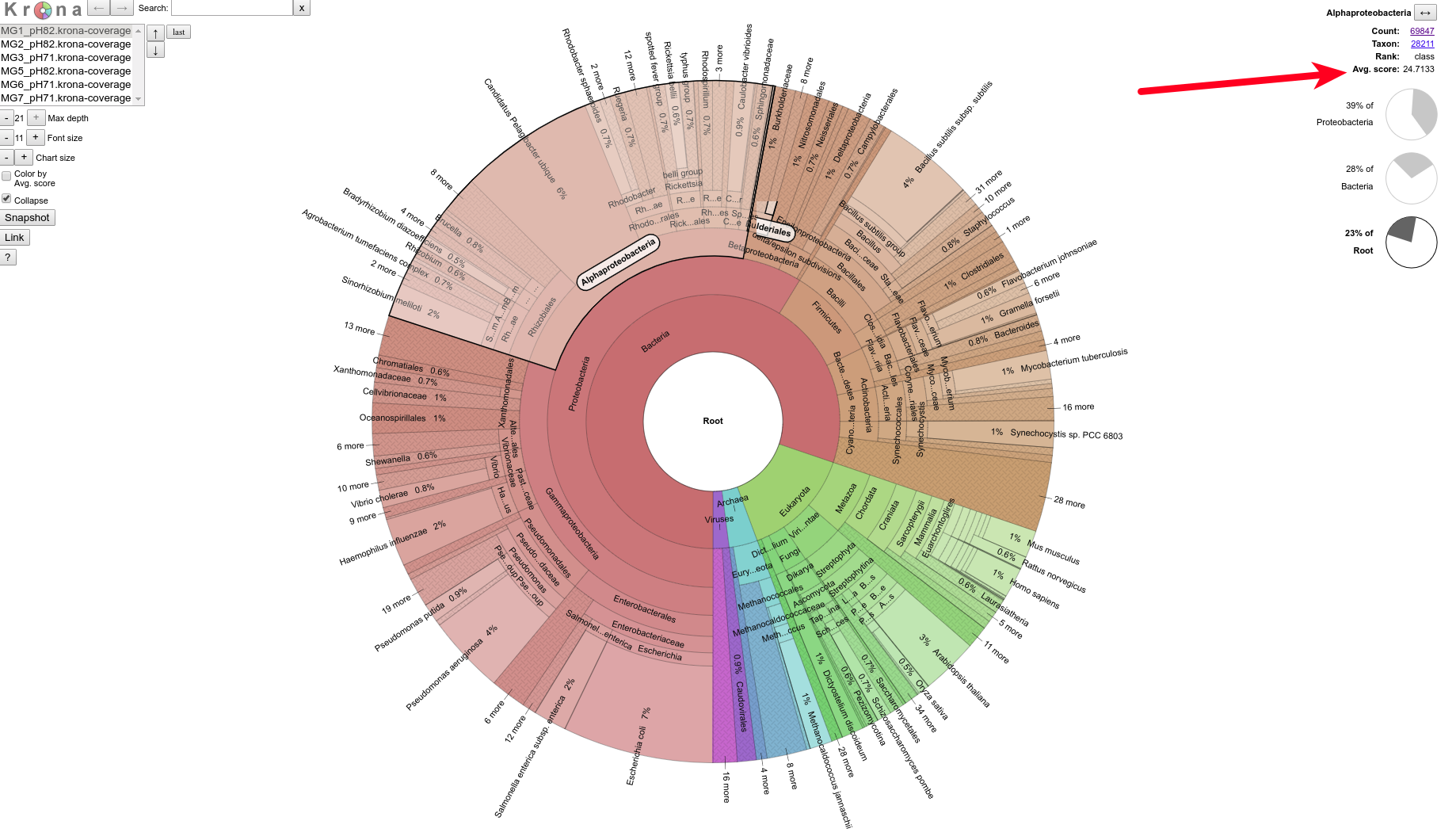
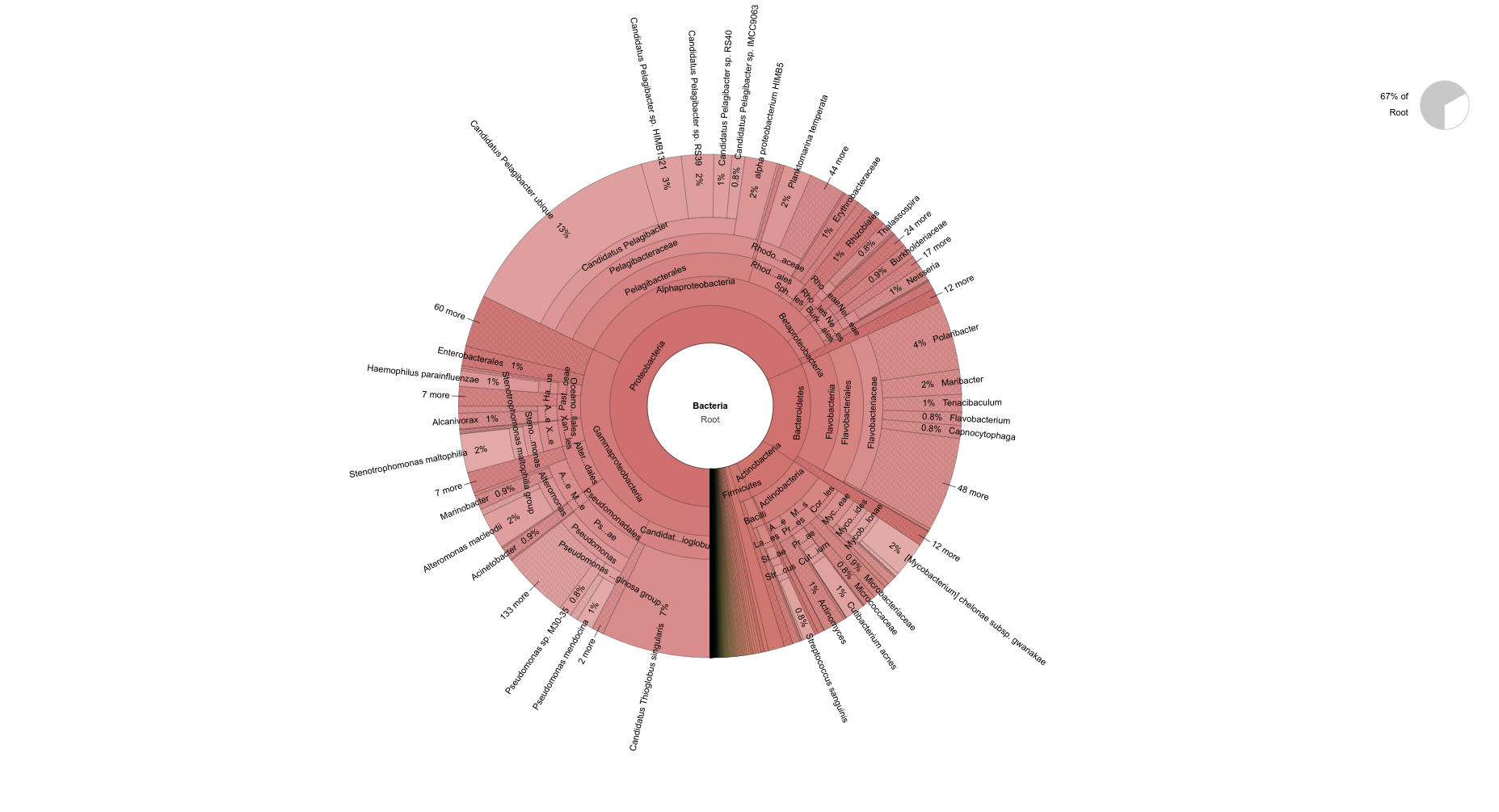
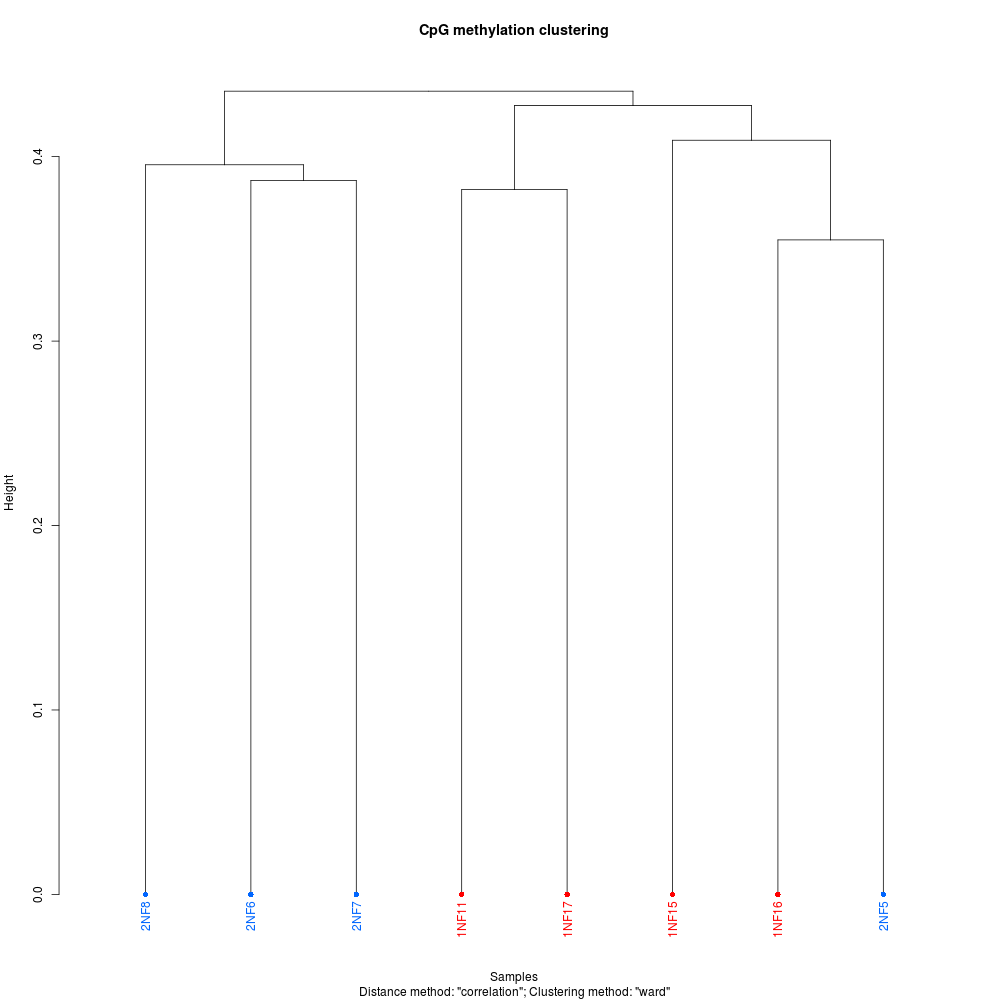
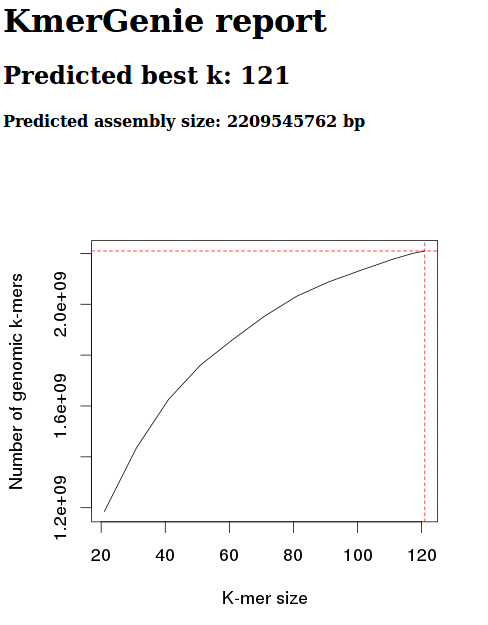
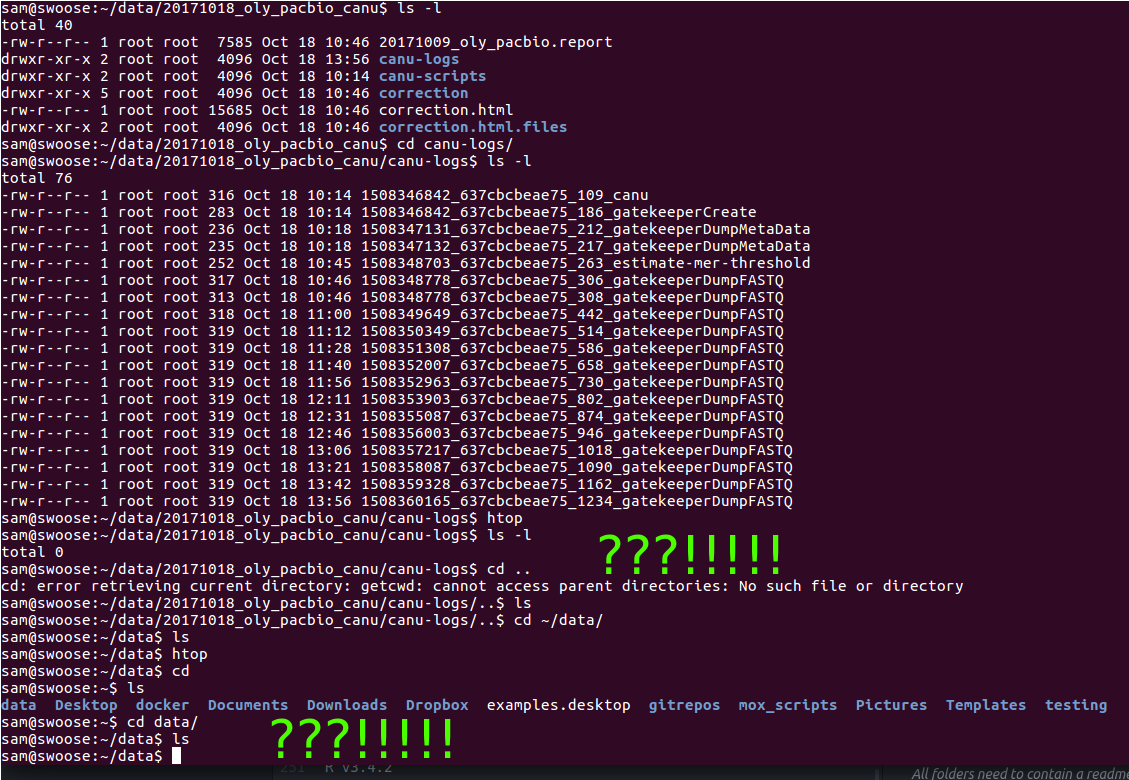
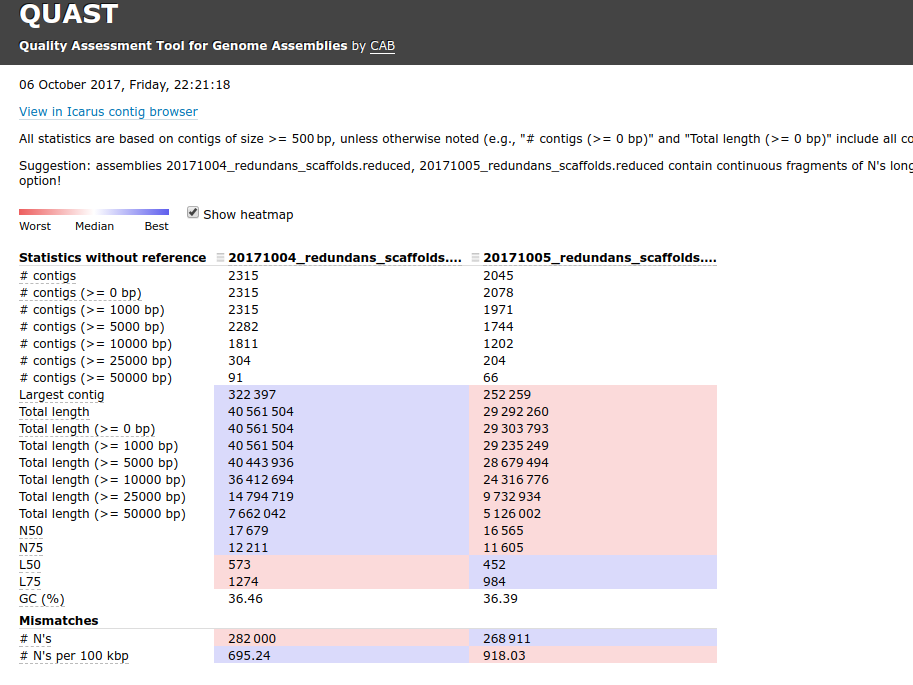
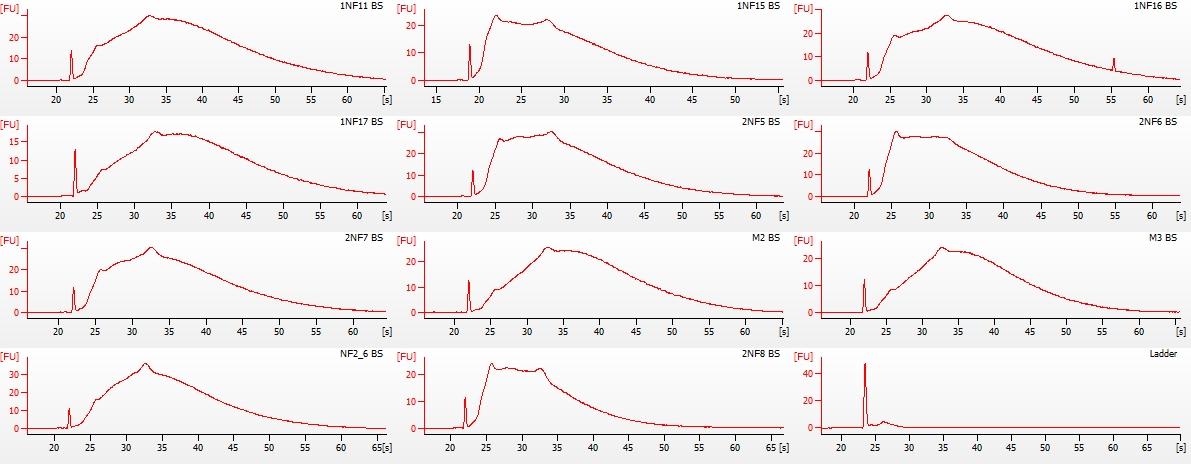
+As part of the CEABIGR project (GitHub repo) Steven performed some inital data wrangling to test out the basic calculations to determine the natural log of the fold change in exon expression, relative to Exon 1, for each gene in a single sample. The decision to perform the calculation in this manner was based on (Li et al. 2018) . I took the next steps to perform this across all samples, as well as generate some comparison plots. After this, will explore how spurious transcription relates to methylation levels across genes.
+See 65-exon-coverage.qmd
+
+
+
+
Code and plots link to commit 8bc64a3.
+
+
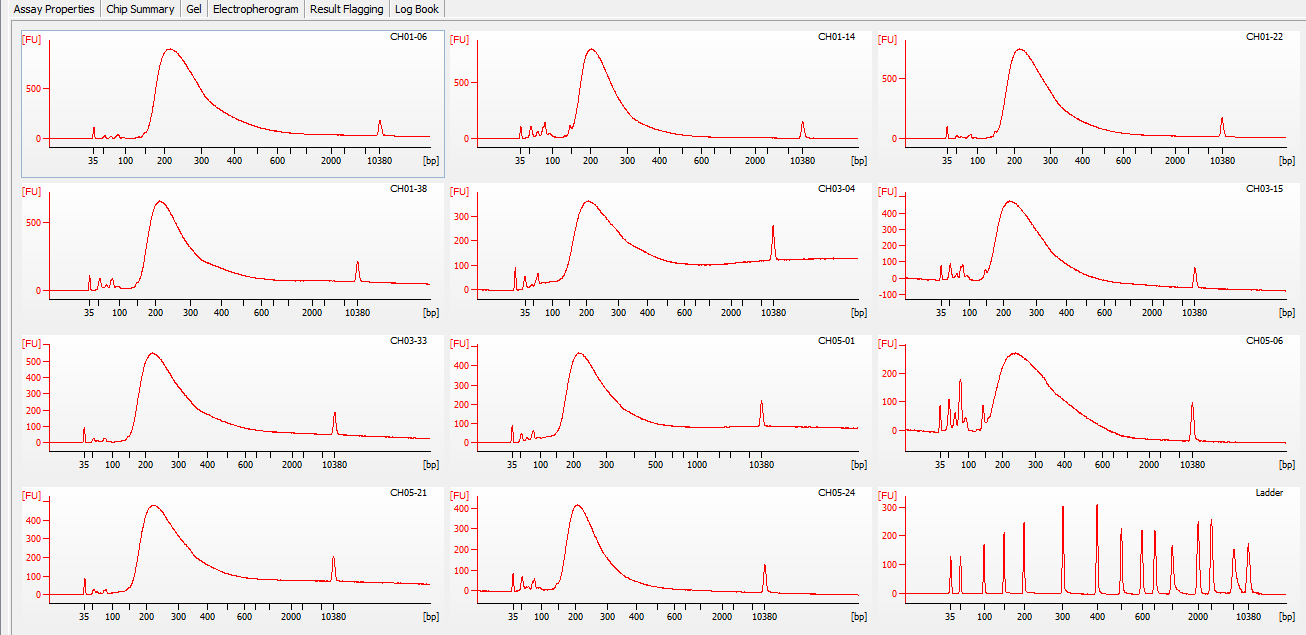
+Plots
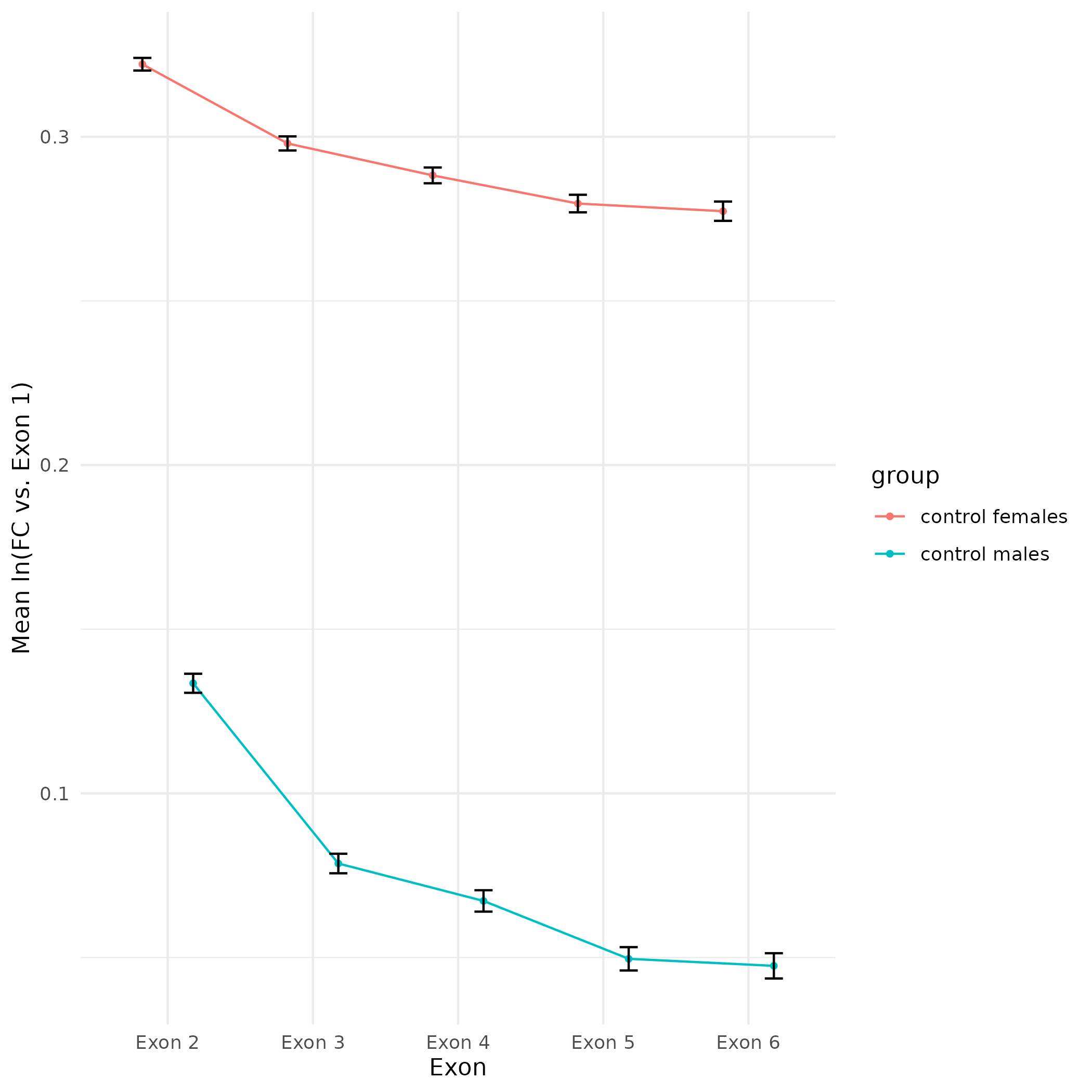
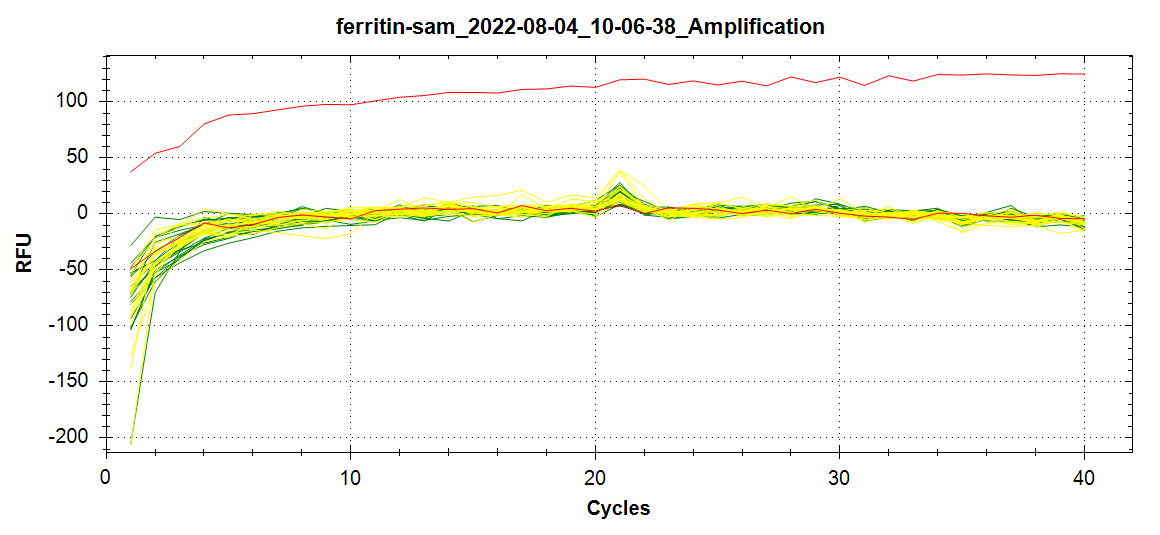
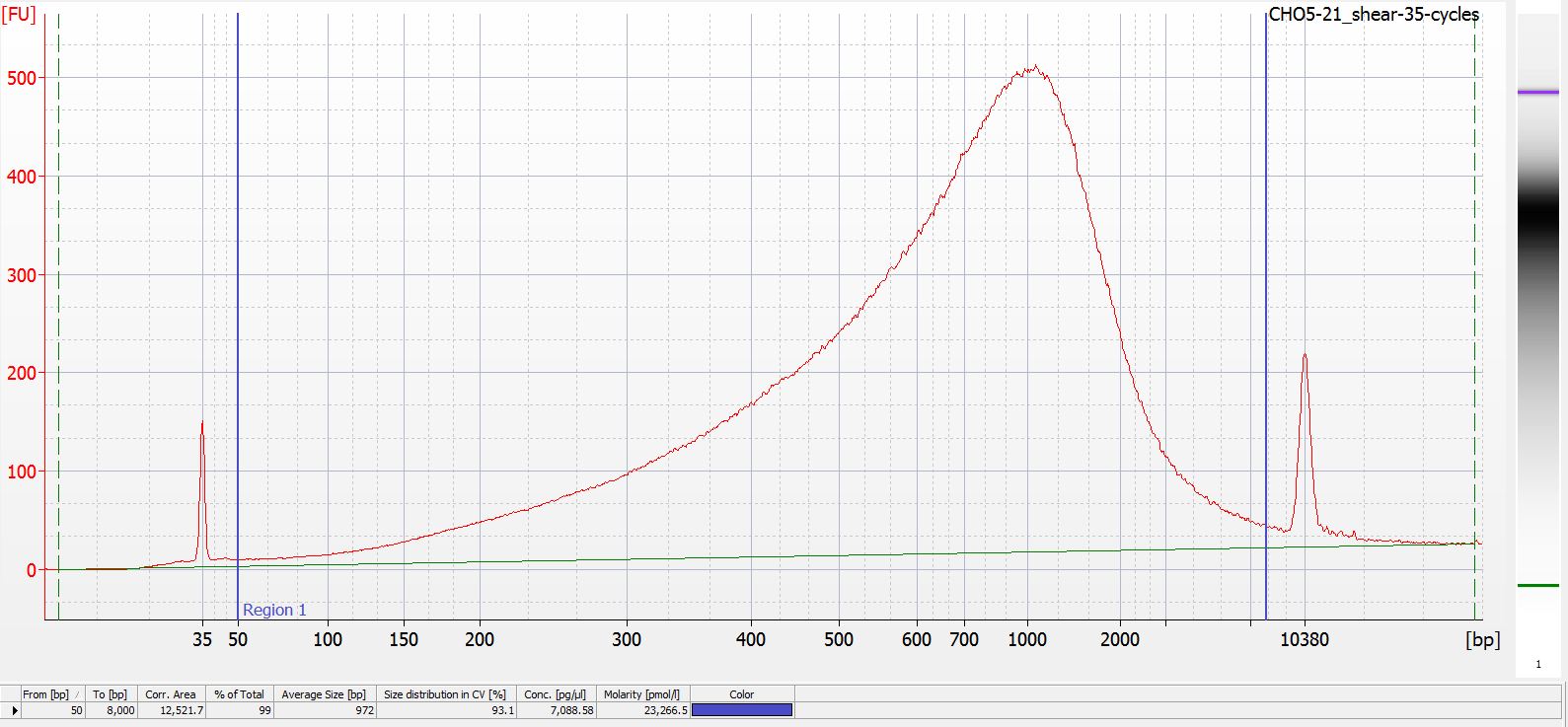
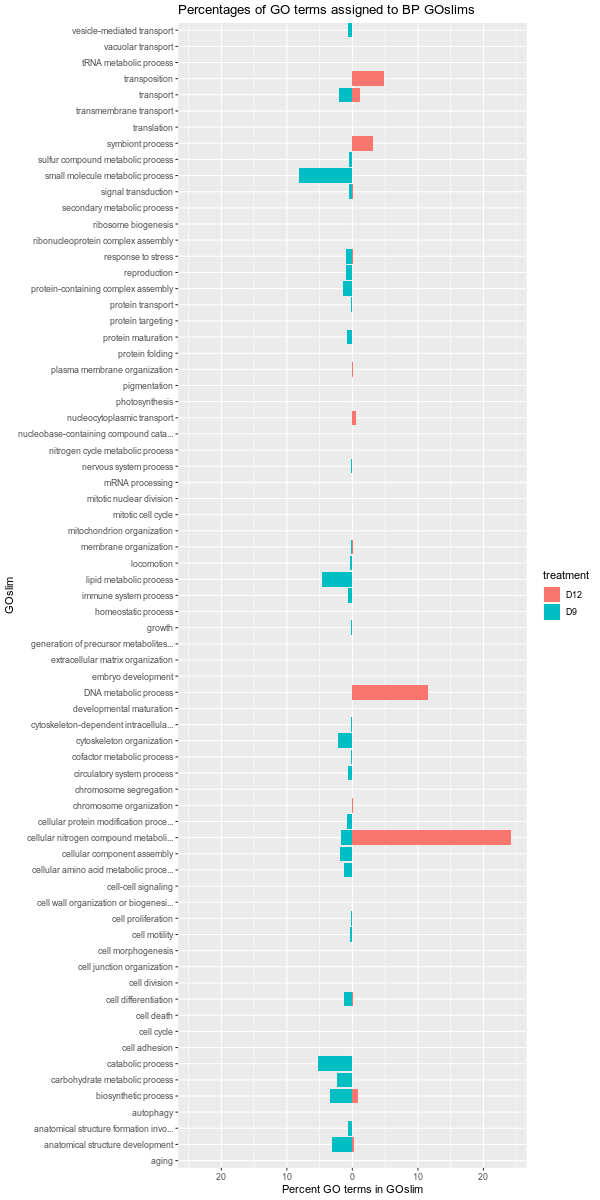
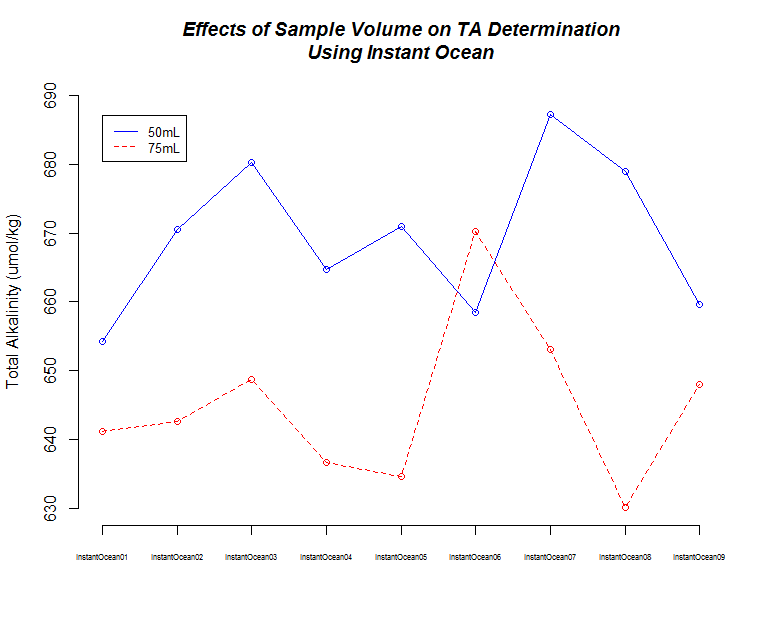
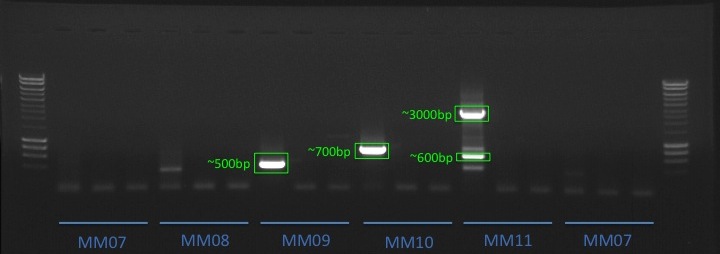
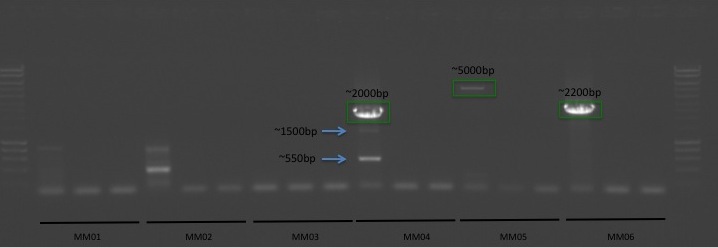
+All plots are line plots of the mean natural log fold-change in exon expression (Exons 2-6), relative to Exon 1. Black bars represent standard error.
+
+
+
+
Plots are simply arranged side-by-side. Scales of axes are not intended to match.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+References
+
+Li, Yong, Yi Jin Liew, Guoxin Cui, Maha J. Cziesielski, Noura Zahran, Craig T. Michell, Christian R. Voolstra, and Manuel Aranda. 2018.
“DNA Methylation Regulates Transcriptional Homeostasis of Algal Endosymbiosis in the Coral Model Aiptasia.” Science Advances 4 (8).
https://doi.org/10.1126/sciadv.aat2142 .
+
+
+ 2024
+ CEABIGR
+ plot
+ Eastern oyster
+ Crassostrea virginica
+ https://robertslab.github.io/sams-notebook/posts/2024/2024-01-03-Data-Exploration---CEABIGR-Spurious-Transcription-Calculations-and-Plotting/index.html
+ Wed, 03 Jan 2024 08:00:00 GMT
+
-
Daily Bits - January 2024
Sam White
@@ -11982,162 +12069,5 @@ font-style: inherit;">>> system_path.log
https://robertslab.github.io/sams-notebook/posts/2023/2023-07-27-DIAMOND-BLASTx---C.virginica-Genes-on-Mox/index.html
Fri, 28 Jul 2023 04:00:00 GMT
-
-
-
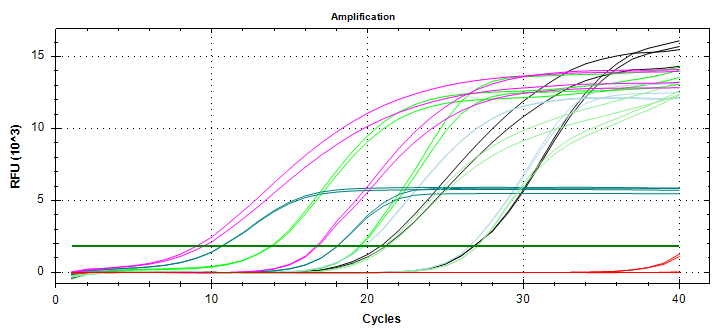
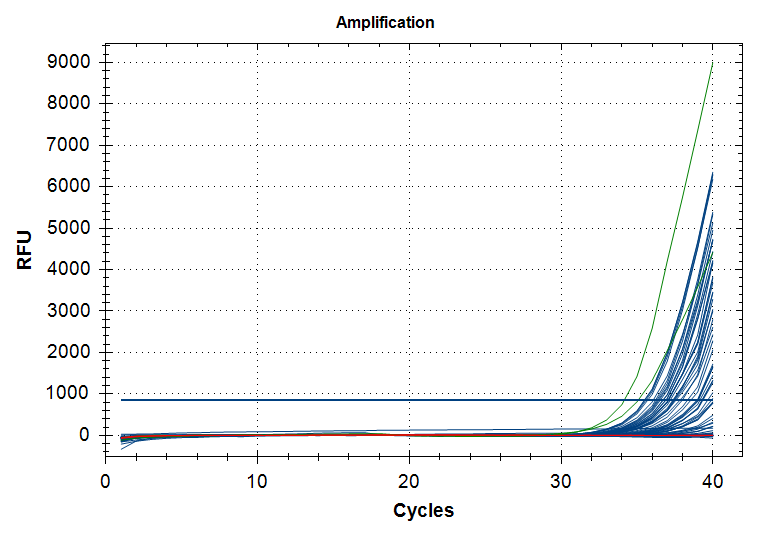
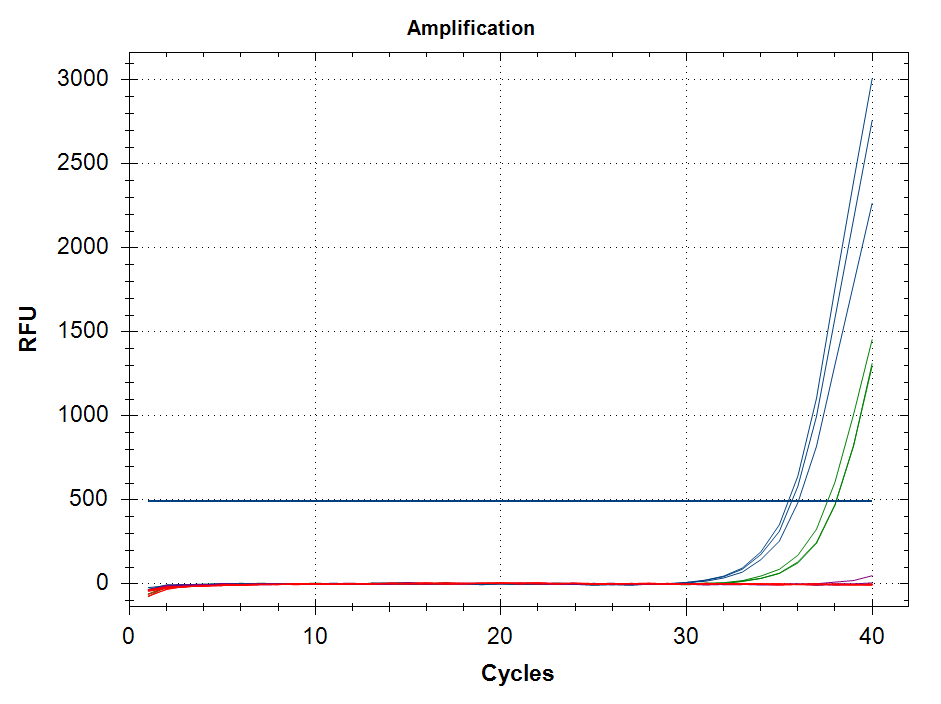
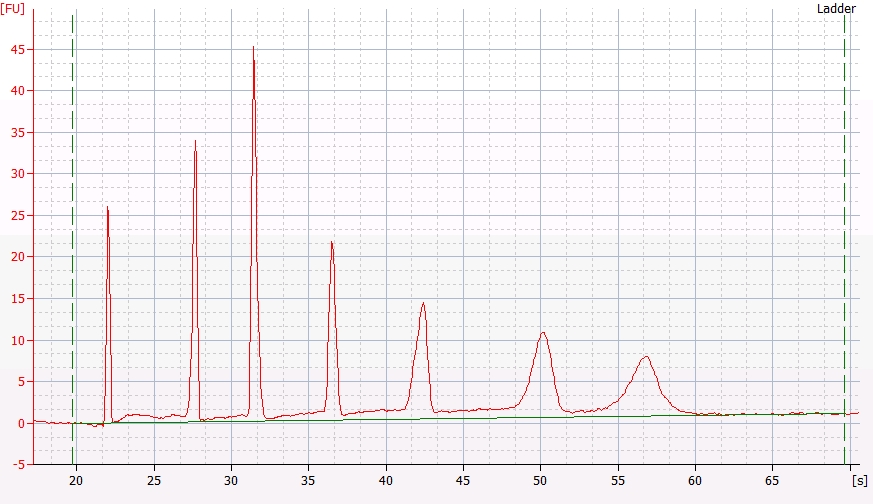
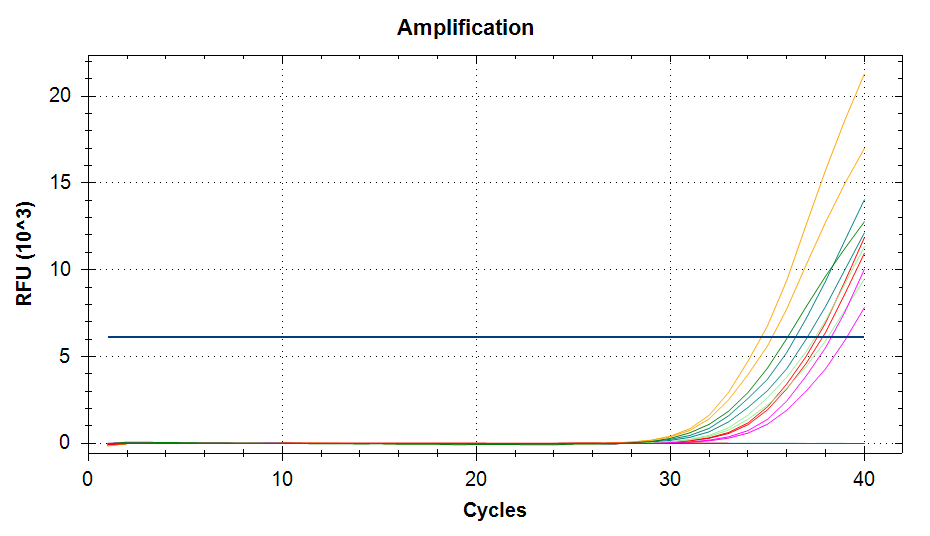
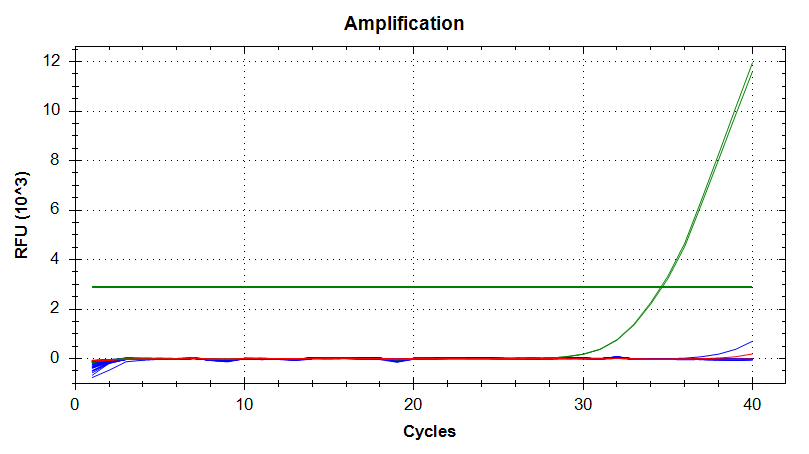
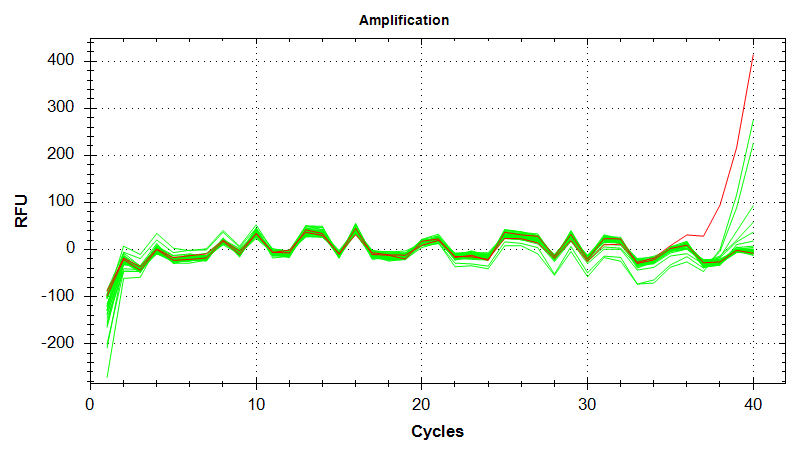
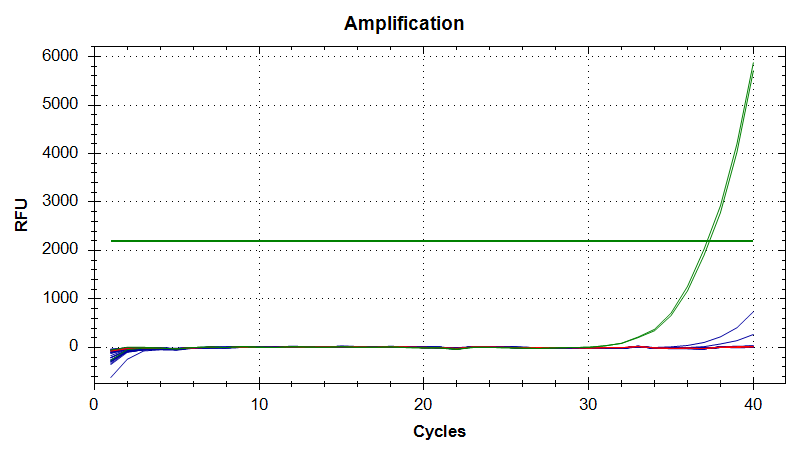
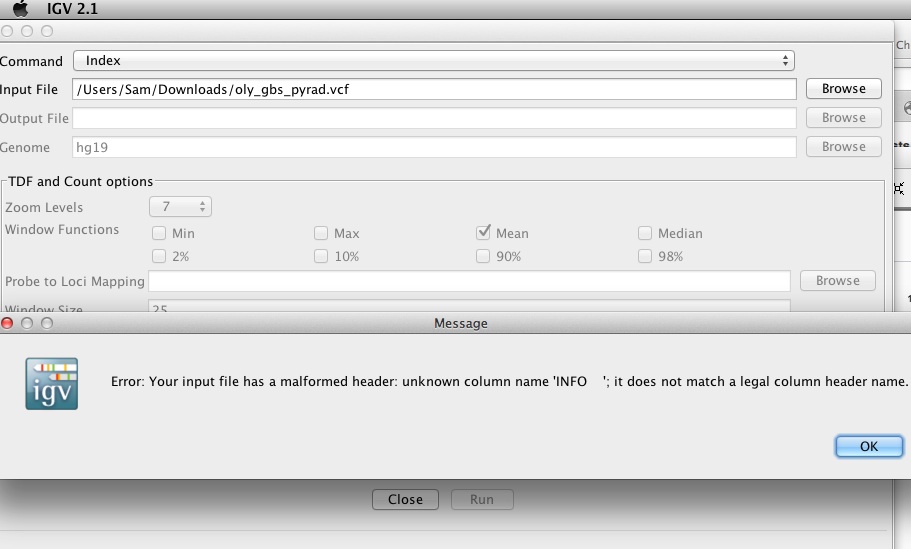
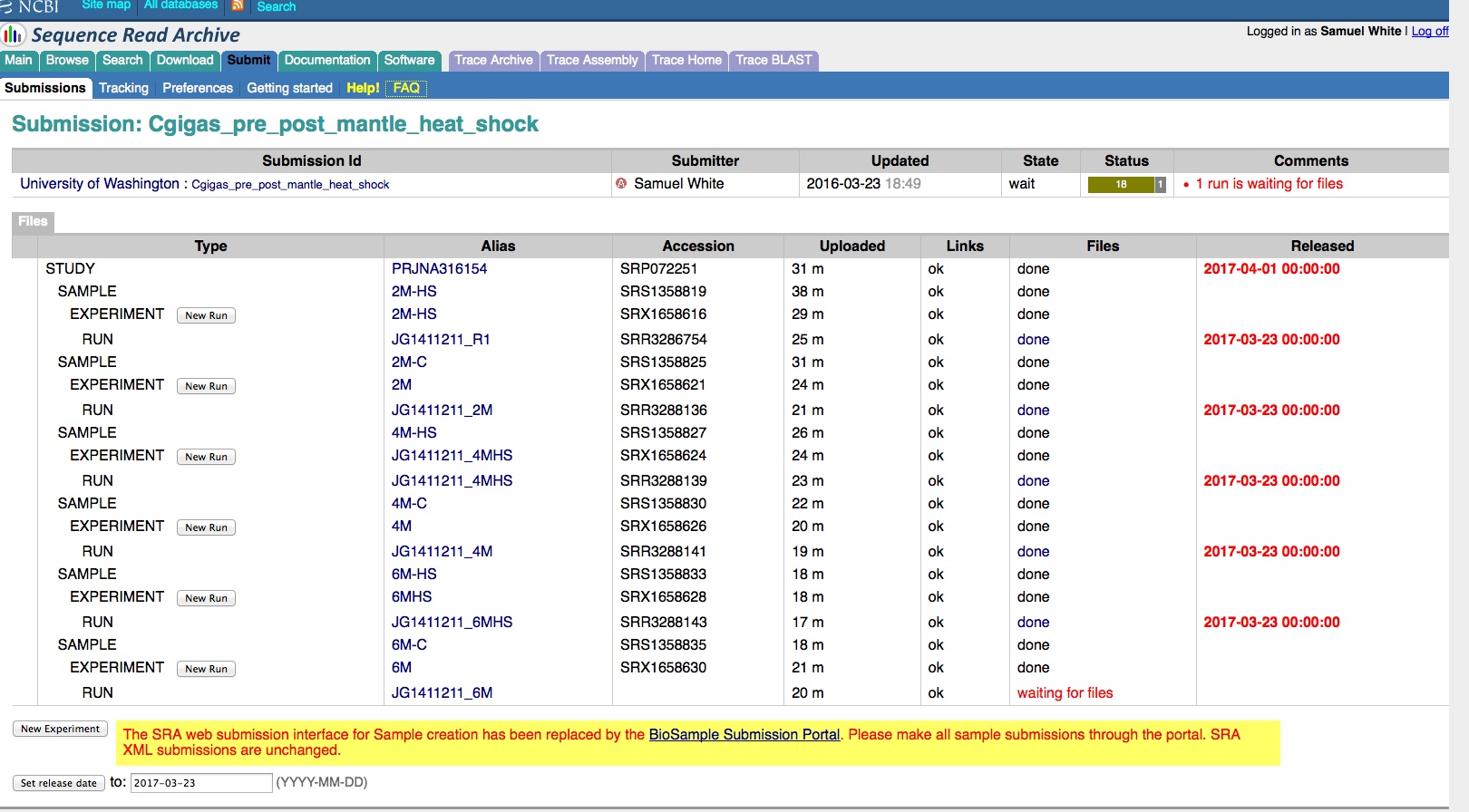
qPCR - C.gigas polyIC
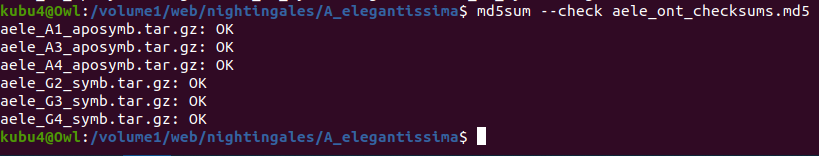
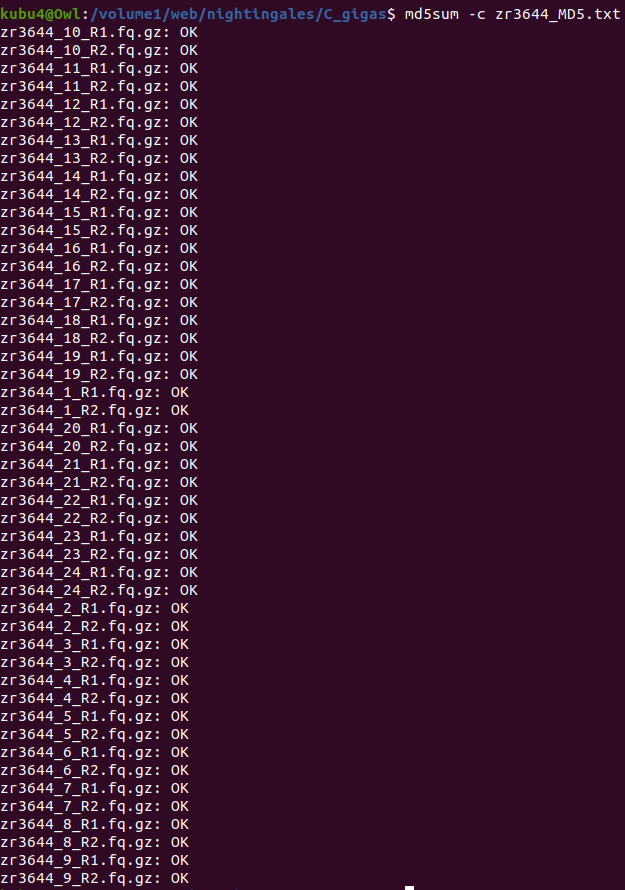
- Sam White
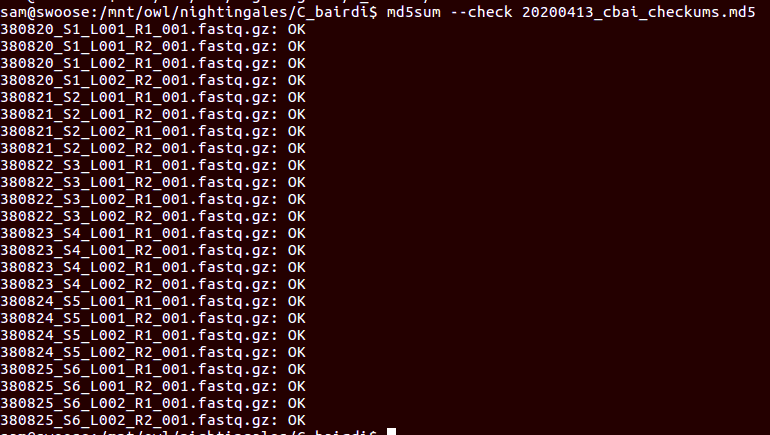
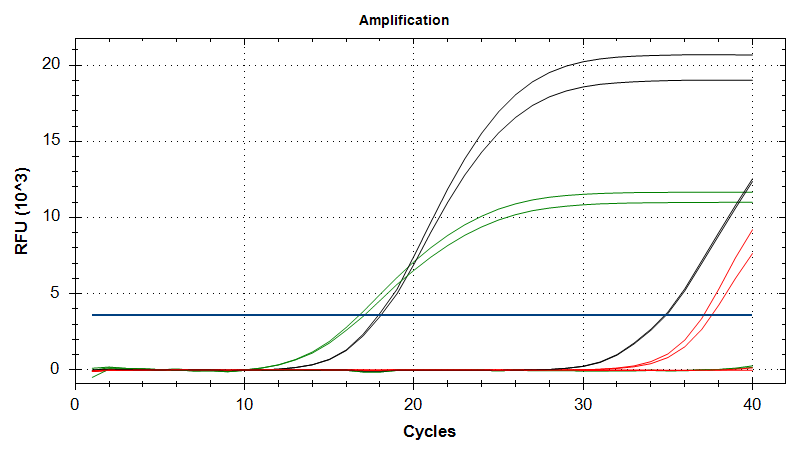
- Using cDNA from 20230721 , performed qPCR on the following primer sets selected by Steven (GitHub Issue), along with Cg_Actin as a potential normalizing gene:
-
-
-
-
-
-
-1170/1
-Cg_Actin
-
-
-1830/1
-Cg_DICER
-
-
-1172/3
-Cg_GAPDH
-
-
-1832/3
-Cg_IRF2
-
-
-1834/5
-Cg_SACSIN
-
-
-1828/9
-Cg_VIPERIN
-
-
-1826/7
-Cg_cGAS
-
-
-1383/4
-Cg_citrate_synthase
-
-
-
-qPCR calcs:
-
-Reactions were run on white, low-profile, 96-well qPCR plates (USA Scientific). A total of two plates were run. One was run on a CFC Connect (BioRad) and the other on a CFX96 (BioRad). See the qPCR Report in the RESULTS section below for cycling params, plate layouts, etc.
-All samples were run in singular . Normally duplicates/triplicates are run. However, due to time and sample constraints, samples were just run in singular . No template controls were run in duplicate.
-
- 2023
- Miscellaneous
- https://robertslab.github.io/sams-notebook/posts/2023/2023-07-26-qPCR---C.gigas-polyIC/index.html
- Wed, 26 Jul 2023 18:42:00 GMT
-
diff --git a/docs/listings.json b/docs/listings.json
index 63100e3ff..a5e1fc81b 100644
--- a/docs/listings.json
+++ b/docs/listings.json
@@ -2,6 +2,7 @@
{
"listing": "/index.html",
"items": [
+ "/posts/2024/2024-01-03-Data-Exploration---CEABIGR-Spurious-Transcription-Calculations-and-Plotting/index.html",
"/posts/2024/2024-01-02-Daily-Bits---January-2024/index.html",
"/posts/2023/2023-12-21-Computer-Management---Returned-Failing-HDDs-from-Gannet-Expansion-Unit/index.html",
"/posts/2023/2023-12-18-Samples-Submitted---Gadus-macrocephalus-liver-tissues-to-Azenta-for-RNA-seq/index.html",
diff --git a/docs/search.json b/docs/search.json
index b2a8833d7..8c551d4ba 100644
--- a/docs/search.json
+++ b/docs/search.json
@@ -11001,7 +11001,7 @@
"href": "index.html",
"title": "University of Washington - SAFS - Roberts Lab",
"section": "",
- "text": "Daily Bits - January 2024\n\n\n\n\n\n\n\n2024\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nJan 2, 2024\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputer Management - Returned Failing HDDs from Gannet Expansion Unit\n\n\n\n\n\n\n\ncomputer management\n\n\n2023\n\n\nHDDs\n\n\ngannet\n\n\n\n\n\n\n\n\n\n\n\nDec 21, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Gadus macrocephalus liver tissues to Azenta for RNA-seq\n\n\n\n\n\n\n\n2023\n\n\nGadus macrocephalus\n\n\nPacific cod\n\n\nAzenta\n\n\nRNA-seq\n\n\nSamples Submitted\n\n\nliver\n\n\nspleen\n\n\ngill\n\n\nblood\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputer Maintenance - Replace Failing HDDs in Synology RX1217 Expansion Unit on Gannet\n\n\n\n\n\n\n\ngannet\n\n\nsynology\n\n\nRX1217\n\n\nHDDs\n\n\n2023\n\n\ncomputer maintenance\n\n\n\n\n\n\n\n\n\n\n\nDec 7, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Investigation - Mysterious Case of Excessive Exons in CEABIGR\n\n\n\n\n\n\n\nCEABIGR\n\n\nCrassostrea viriginica\n\n\nEastern oyster\n\n\n2023\n\n\n\n\n\n\n\n\n\n\n\nDec 6, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - December 2023\n\n\n\n\n\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPlotting Fun - Lab Meeting Task\n\n\n\n\n\n\n\nplotting\n\n\nR\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - M.magister De Novo Transcriptome Assembly Using Trinotate on Mox\n\n\n\n\n\n\n\nmox\n\n\n2023\n\n\nMetacarcinus magister\n\n\nDungenss crab\n\n\nTrinotate\n\n\ntranscriptome\n\n\nannotation\n\n\n\n\n\n\n\n\n\n\n\nNov 9, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - M.magister De Novo Transcriptome Assembly for DuMOAR Project Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\nmox\n\n\nDIAMOND\n\n\nBLASTx\n\n\ntranscriptome\n\n\n2023\n\n\nDuMOAR\n\n\nDungeness crab\n\n\nMetacarcinus magister\n\n\n\n\n\n\n\n\n\n\n\nNov 9, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - November 2023\n\n\n\n\n\n\n\nDaily Bits\n\n\n2023\n\n\n\n\n\n\n\n\n\n\n\nNov 5, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR Analysis - C.gigas Matt George Poly:IC Diploids\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 22, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Identification and Alignments - C.virginica RNAseq with NCBI Genome GCF_002022765.2 Using Hisat2 and Stringtie on Mox Again\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 21, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas PolyIC Diploid MgCl2\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 17, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - C.gigas PolyIC Diploid MgCl2\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Quantification - C.gigas PolyIC Diploid MgCl2\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - August 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nAug 1, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nsRNA-seq Alignments - E5 Coral P.evermanni Using ShortStack on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Extractions - M.magister MEGAN FastA to FastQ Arthropoda and Unassigned Reads\n\n\n\n\n\n\n\n2023\n\n\nDuMOAR\n\n\n\n\n\n\n\n\n\n\n\nJul 30, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDIAMOND BLASTx - C.virginica Genes on Mox\n\n\n\n\n\n\n\n2023\n\n\nCEABIGR\n\n\n\n\n\n\n\n\n\n\n\nJul 27, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas polyIC\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.virginica Genes Only FastA from Genes BED File Using gffread on Raven\n\n\n\n\n\n\n\n2023\n\n\nCEABIGR\n\n\n\n\n\n\n\n\n\n\n\nJul 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Extractions - M.magister MEGAN Arthropoda and Unassigned Reads to FastA\n\n\n\n\n\n\n\n2023\n\n\nDuMOAR\n\n\n\n\n\n\n\n\n\n\n\nJul 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Quantification - C.gigas PolyIC\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 21, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - C.gigas PolyIC RNA\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 21, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas Ctenidia cDNA for Noah’s Heat-Mechanical Stress Project\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 21, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas Ctenidia cDNA for Noah’s Heat-Mechanical Stress Project\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 19, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas cDNA Primer Tests for Noah’s Heat-Mechanical Stress Project\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 13, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - C.gigas RNA from Noah’s Heat-Mechanical Stress Project\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 13, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Quantification - C.gigas RNA from Noah’s Heat-Mechanical Stress Project\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 13, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - C.gigas Ctenidia from Noah’s Heat-Mechanical Stress Project\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 12, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - July 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nJul 5, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFile Conversion - M.magister MEGANized DAA to RMA6\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Pacific Cod (G.macrocephalus) Sequencing Data from Novogene\n\n\n\n\n\n\n\n2023\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nsRNA-seq Alignments - E5 Coral A.pulchra P.meandrina Using ShortStack on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nJun 28, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming and QC - E5 Coral sRNA-seq Data fro A.pulchra P.evermanni and P.meandrina Using FastQC flexbar and MultiQC on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nJun 20, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nORF Identification - L.staminea De Novo Transcriptome Assembly v1.0 Using Transdecoder on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 17, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - De Novo L.staminea Trimmed RNAseq Using Trinity on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - L.staminea RNA-seq Using FastQC fastp and MultiQC on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - June 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nJun 2, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeats Identification - P.meandrina Using RepeatMasker on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nMay 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming and QC - E5 Coral sRNA-seq Trimming Parameter Tests and Comparisons\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nMay 24, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - P.meandrina Genome GFF to GTF Using gffread\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQ QC and Trimming - E5 Coral RNA-seq Data for A.pulchra P.evermanni and P.meandrina Using FastQC fastp and MultiQC on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - E5 Coral RNA-seq and sRNA-seq Reorganizing and Renaming\n\n\n\n\n\n\n\n2023\n\n\nData Management\n\n\n\n\n\n\n\n\n\n\n\nMay 17, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Coral RNA-seq Data from Azenta Project 30-789513166\n\n\n\n\n\n\n\n2023\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Coral sRNA-seq Data from Azenta Project 30-852430235\n\n\n\n\n\n\n\n2023\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nMay 15, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nlncRNA Expression - P.generosa lncRNA Expression Using StringTie\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 4, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - May 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nMay 2, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nlncRNA Identification - P.generosa lncRNAs using CPC2 and bedtools\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 2, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nContainers - Apptainer Explorations\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 28, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Alignments - P.generosa RNA-seq Alignments for lncRNA Identification Using Hisat2 StingTie and gffcompare on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQ Trimming and QC - P.generosa RNA-seq Data from 20220323 on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - CEABIGR C.virginica Exon Expression Table\n\n\n\n\n\n\n\n2023\n\n\nCEABIGR\n\n\n\n\n\n\n\n\n\n\n\n\nApr 5, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - April 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nApr 3, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Append Gene Ontology Aspect to P.generosa Primary Annotation File\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 29, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing Read Taxonomic Classification - M.magister RNA-seq Using DIAMOND BLASTx and MEGAN6 daa-meganizer on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - March 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nMar 3, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Trimmed M.magister RNA-seq from NOAA\n\n\n\n\n\n\n\n2023\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing Read Taxonomic Classification - P.verrucosa E5 RNA-seq Using DIAMOND BLASTx and MEGAN daa-meganizer on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nFeb 21, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.goreaui Genome GFF to GTF Using gffread\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nFeb 17, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Identification and Alignments - P.verrucosa RNA-seq with Pver_genome_assembly_v1.0 Using HiSat2 and Stringtie on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nFeb 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQ Trimming and QC - P.verrucosa RNA-seq Data from Danielle Becker in Hollie Putnam Lab Using fastp FastQC and MultiQC on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nFeb 15, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Create P.verrucosa GCA_014529365.1 Karyotype File\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nFeb 15, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - P.verrucosa RNA-seq and WGBS Full Data from Danielle Becker\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nFeb 10, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - February 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nFeb 1, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Indexing - P.verrucosa v1.0 Assembly with HiSat2 on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 31, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Indexing - M.capitata HIv3 Assembly with HiSat2 on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 31, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Indexing - P.acuta HIv2 Assembly with HiSat2 on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 31, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - P.verrucosa Genome GFF to GTF Using gffread\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 27, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - M.capitata Genome GFF to GTF Using gffread\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 27, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - P.acuta Genome GFF to GTF Conversion Using gffread\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Indexing - P.verrucosa NCBI GCA_014529365.1 with HiSat2 on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 25, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Indexing - M.capitata NCBI GCA_006542545.1 with HiSat2 on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 25, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Data - Coral SRA BioProject PRJNA744403 Download and QC\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 13, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMonthly Goals - January 2023\n\n\n\n\n\n\n\n2023\n\n\nMonthly Goals\n\n\n\n\n\n\n\n\n\n\n\nJan 5, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - January 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nJan 3, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBS-Seq Analysis - Nextflow EpiDiverse SNP Pipeline for Haws Hawaii C.gigas BAMs from Yaamini Base Config\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 15, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBS-Seq Analysis - Nextflow EpiDiverse SNP Pipeline for Haws Hawaii C.gigas BAMs from Yaamini\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 14, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - December 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - November 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nNov 1, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - October 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nOct 1, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.virginica NCBI GCF_002022765.2 GFF to Gene and Pseudogene Combined BED File\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 26, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBSseq SNP Analysis - Nextflow EpiDiverse SNP Pipeline for C.virginica CEABIGR BSseq data\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 21, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Identify C.virginica Genes with Different Predominant Isoforms for CEABIGR\n\n\n\n\n\n\n\n2022\n\n\nCEABIGR\n\n\n\n\n\n\n\n\n\n\n\nSep 20, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Alignments - P.generosa Alignments and Alternative Transcript Identification Using Hisat2 and StringTie on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 14, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQ Trimming - Geoduck RNAseq Data Using fastp on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 8, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - September 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nSep 2, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQ Trimming and QC - C.virginica Larval BS-seq Data from Lotterhos Lab and Part of CEABIGR Project Using fastp on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 29, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Convert S.namaycush NCBI GFF to genes-only BED file for Use in Ballgown Analysis\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 18, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSplice Site Identification - S.namaycush Liver Parasitized and Non-Parasitized SRA RNAseq Using Hisat2-Stingtie with Genome GCF_016432855.1\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 10, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Repeat of Mussel Gill Heat Stress cDNA with Ferritin Primers\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 4, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - August 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nAug 3, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Dorothys Mussel cDNA from 20220726\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 27, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - Dorothy Mussel Gill Samples\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 26, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBS-seq and SNP Analysis - Nextflow EpiDiverse Pipelines Trials and Tribulations\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 25, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Data - S.namaycush SRA BioProject PRJNA674328 Download and QC\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - July 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nJul 5, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - June 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nJun 22, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Ostrea lurida MBD BS-seq from 20160203\n\n\n\n\n\n\n\n2022\n\n\nSRA Submission\n\n\n\n\n\n\n\n\n\n\n\nJun 15, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nProject Summary - C.virginica CEABiGR - Female vs. Male Gonad Exposed to OA\n\n\n\n\n\n\n\n2022\n\n\nProject Summary\n\n\n\n\n\n\n\n\n\n\n\nApr 25, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Create Primary P.generosa Genome Annotation File\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 19, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - O.nerka Berdahl Brain Tissues\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 11, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nServer Maintenance - Fix Server Certificate Authentication Issues\n\n\n\n\n\n\n\n2022\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nApr 2, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNextflow - Trials and Tribulations of Installing and Using NF-Core RNAseq\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 25, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - P.generosa Genomic Feature FastA Creation\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 24, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDifferential Gene Expression - P.generosa DGE Between Tissues Using Nextlow NF-Core RNAseq Pipeline on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 23, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - C.virginica BSseq Unmapped Reads Using MEGAN6\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 9, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - C.virginica RNAseq Zymo ZR4059 Analyzed by ZymoResearch\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTaxonomic Assignment - C.virginica BSseq Unmapped Reads Using DIAMOND BLASTx and MEGAN6 on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - P.generosa Genome GFF Conversion to GTF Using gffread\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Identification and Alignments - C.virginica RNAseq with NCBI Genome GCF_002022765.2 Using Hisat2 and Stringtie on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 25, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - Additional 20bp from C.virginica Gonad RNAseq with fastp on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 24, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.virginica lncRNA Extractions from NCBI GCF_002022765.2 Using GffRead\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 17, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Genome-guided C.virginica Adult Gonad OA RNAseq Using Trinity on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Alignment - C.virginica Adult OA Gonad Data to GCF_002022765.2 Genome Using HISAT2 on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 31, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.virginica Gonad RNAseq Transcript Counts Per Gene Per Sample Using Ballgown\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 27, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.virginica OA Larvae DNA Methylation FastQs from Lotterhos Lab\n\n\n\n\n\n\n\n2022\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nJan 19, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nProject Summary - Matt George PSMFC Mytilus Byssus Project\n\n\n\n\n\n\n\n2022\n\n\nProject Summary\n\n\n\n\n\n\n\n\n\n\n\nJan 13, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - O.nerka Berdahl Brain Tissues\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - M.trossulus Gill\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - M.trossulus Gill and Phenol Gland\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - M.trossulus Gill and Phenol Gland\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 10, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - M.trossulus Gill and Phenol Gland\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - M.trossulus Phenol Gland and Gill\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 23, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - M.trossulus Foot and Phenol Gland\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nProject Summary - O.nerka Berdahl Samples\n\n\n\n\n\n\n\n2021\n\n\nProject Summary\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - O.nerka Berdahl Brain Tissues\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - O.nerka Berdahl Tissues\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 20, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.virginica NCBI GCF_002022765.2 GFF to Gene BED File\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 9, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDifferential Transcript Expression - C.virginica Gonad RNAseq Using Ballgown\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 21, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSNP Characterization - C.bairdi v3.1 Transcriptome Assembly and Day2 DEG Pooled Samples RNAseq Data\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSNP Identification - C.bairdi Day 2 DEG Pooled Samples Using bcftools on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nSep 9, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Alignments - C.bairdi Day 2 Infected-Uninfected Temperature Increase-Decrease RNAseq to cbai_transcriptome_v3.1.fasta with Hisat2 on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nSep 8, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Indexing - C.bairdi Transcriptome cbai_transcriptome_v3.1.fasta with Hisat2 on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nSep 8, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputer Servicing - APC SUA2200RM2U UPS Battery Replacement\n\n\n\n\n\n\n\n2021\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nAug 24, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputer Management - Disable Sleep and Hibernation on Raven\n\n\n\n\n\n\n\n2021\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nAug 24, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nUninterruptible Power Supply - Battery Replacement for APS BR1000G\n\n\n\n\n\n\n\n2021\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nAug 11, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Identification and Quantification - C.virginia RNAseq With NCBI Genome GCF_002022765.2 Using StringTie on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 26, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotations - Splice Site and Exon Extractions for C.virginica GCF_002022765.2 Genome Using Hisat2 on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 20, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - C.virginica Gonad RNAseq with FastP on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSummary - Geoduck RNAseq Data\n\n\n\n\n\n\n\n2021\n\n\nProject Summary\n\n\n\n\n\n\n\n\n\n\n\nJul 12, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Analysis - Identification of Potential Contaminating Sequences in Panopea-generosa-v1.0 Assembly Using BlobToolKit on Mox\n\n\n\n\n\n\n\n2021\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 12, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQC - Yaamini’s C.virginica RNAseq and WGBS from ZymoResearch on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 29, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - S.salar Gene Annotations from NCBI RefSeq GCF_000233375.1_ICSASG_v2_genomic.gff for Shelly\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Yaamini’s C.virginica WGBS and RNAseq Data from ZymoResearch\n\n\n\n\n\n\n\n2021\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nMay 28, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Comparisons - Ostrea lurida Non-scaffold Genome Assembly Comparisons Using Quast on Swoose\n\n\n\n\n\n\n\n2021\n\n\nOlympia Oyster Genome Assembly\n\n\n\n\n\n\n\n\n\n\n\nMay 21, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Assessment - Olurida_v090 Using Quast on Swoose\n\n\n\n\n\n\n\n2021\n\n\nOlympia Oyster Genome Assembly\n\n\n\n\n\n\n\n\n\n\n\nMay 21, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olurida_v090 with BGI Illumina and PacBio Hybrid Using Wengan on Mox\n\n\n\n\n\n\n\n2021\n\n\nOlympia Oyster Genome Assembly\n\n\n\n\n\n\n\n\n\n\n\nMay 20, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - O.lurida BGI FastQs with FastP on Mox\n\n\n\n\n\n\n\n2021\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 18, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Submission - Validation of Olurida_v081.fa and Annotated GFFs Prior to Submission to NCBI\n\n\n\n\n\n\n\n2021\n\n\nOlympia Oyster Genome Assembly\n\n\n\n\n\n\n\n\n\n\n\nMay 13, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeatMasker - C.gigas Rosling NCBI Genome GCA_902806645.1 on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 4, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSingularity - RStudio Server Container on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 23, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Mapping - 10x-Genomics Trimmed FastQ Mapped to P.generosa v1.0 Assembly Using Minimap2 for BlobToolKit on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - P.generosa v1.0 Assembly Using DIAMOND BLASTx for BlobToolKit on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - P.generosa v1.0 Assembly Using BLASTn for BlobToolKit on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming P.generosa 10x Genomics HiC FastQs with fastp on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 1, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate on C.bairdi Transcriptome v4.0 on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - DIAMOND BLASTx on C.bairdi Transcriptome v4.0 on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - C.bairdi Transcriptome v4.0 on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 17, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v4.0 on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 17, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi Transcriptome v4.0 Using Trinity on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 17, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Extractions - C.bairdi RNAseq Reads from C.opilio BLASTx Matches with seqkit on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 16, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDIAMOND BLASTx - C.bairdi RNAseq vs C.opilio Genome Proteins on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 12, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate Hematodinium v1.7 on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 9, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate Hematodinium v1.6 on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 9, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Hematodinium Transcriptomes v1.6 and v1.7 with Trinity on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 8, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Gene ID Extraction from P.generosa Genome GFF Using Methylation Machinery Gene IDs\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 26, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Gene ID Extraction from P.generosa Genome GFF Using Methylation Machinery List\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 26, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - A.elegantissima ONT Fast5 from Jay Dimond\n\n\n\n\n\n\n\n2021\n\n\nSRA Submission\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Anthopleura elegantissima - aggregating anenome - NanoPore Genome Sequence from Jay Dimond\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 1, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - M.magister MBD-BSseq Libraries to Univ. of Oregon GC3F\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Comparisons - C.bairdi Transcriptomes Evaluations with DETONATE rsem-eval on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 29, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAlignments - C.bairdi RNAseq Transcriptome Alignments Using Bowtie2 on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 24, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Cockle Clam Gonad H and E Slides\n\n\n\n\n\n\n\n2020\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQC - M.magister MBD-BSseq Pool Test MiSeq Run on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - M.magister MBD-BSseq Pool Test MiSeq Run\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAlignment - C.gigas RNAseq to GCF_000297895.1_oyster_v9 Genome Using STAR on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Haws Lab C.gigas Ploidy pH WGBS\n\n\n\n\n\n\n\n2020\n\n\nSRA Submission\n\n\n\n\n\n\n\n\n\n\n\nDec 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - Haws Lab C.gigas Ploidy pH WGBS 10bp 5 and 3 Prime Ends Using fastp and MultiQC on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 6, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQc - C.gigas Ploidy pH WGBS Raw Sequence Data from Haws Lab on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 6, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.gigas Diploid-Triploid pH Treatments Ctenidia WGBS from ZymoResearch\n\n\n\n\n\n\n\n2020\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nDec 5, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - Ronits C.gigas Ploidy WGBS 10bp 5 and 3 Prime Ends Using fastp and MultiQC on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - M.magister MBD BSseq Libraries for MiSeq at NOAA\n\n\n\n\n\n\n\n2020\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Quantification - M.magister MBD BSseq Libraries with Qubit\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Cockle Clam Gonad Histology Cassettes for H and E\n\n\n\n\n\n\n\n2020\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - Ronits C.gigas Ploidy WGBS Using fastp and MultiQC on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - M.magister MBD BSseq Libraries\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD BSseq Library Prep - M.magister MBD-selected DNA Using Pico Methyl-Seq Kit\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - P.generosa Hemocytes from Shelly\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Ronits C.gigas Ploidy WGBS\n\n\n\n\n\n\n\n2020\n\n\nSRA Submission\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQc - C.gigas Ploidy WGBS Raw Sequence Data from Ronits Project on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - Crustacean Transcripome Completeness Evaluation Using BUSCO on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.gigas Ploidy WGBS from Ronits Project via ZymoResearch\n\n\n\n\n\n\n\n2020\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nNov 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - MultiQC on S.salar RNAseq from fastp and HISAT2 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 6, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHard Drive Upgrade - Gannet Synology Server\n\n\n\n\n\n\n\n2020\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nNov 5, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Alignments - S.salar HISAT2 BAMs to GCF_000233375.1_ICSASG_v2_genomic.gtf Transcriptome Using StringTie on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 4, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Alignments - Trimmed S.salar RNAseq to GCF_000233375.1_ICSASG_v2_genomic.fa Using Hisat2 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 3, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Selection - M.magister Sheared Gill gDNA 16 of 24 Samples Set 3 of 3\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 3, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Selection - M.magister Sheared Gill gDNA 8 of 24 Samples Set 2 of 3\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - Shelly S.salar RNAseq Using fastp and MultiQC on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 29, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Selection - M.magister Sheared Gill gDNA 8 of 24 Samples Set 1 of 3\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing - M.magister gDNA Additional Shearing CH05-01_21 CH07-11 and Bioanalyzer\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing - M.magister gDNA Shearing All Samples and Bioanalyzer\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing - M.magister CH05-21 gDNA Full Shearing Test and Bioanalyzer\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing - M.magister gDNA Shear Testing and Bioanalyzer\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 21, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Mapping - C.bairdi 201002558-2729-Q7 and 6129-403-26-Q7 Taxa-Specific NanoPore Reads to cbai_genome_v1.01.fasta Using Minimap2 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.bairdi NanoPore Reads Extractions With Seqtk on Mephisto\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 13, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoPore Reads Extractions - C.bairdi Taxonomic Reads Extractions with MEGAN6 on 201002558-2729-Q7 and 6129-403-26-Q7\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComparison - C.bairdi 20102558-2729 vs. 6129-403-26 NanoPore Taxonomic Assignments Using MEGAN6\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTaxonomic Assignments - C.bairdi 6129-403-26-Q7 NanoPore Reads Using DIAMOND BLASTx on Mox and MEGAN6 daa2rma on emu\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTaxonomic Assignments - C.bairdi 20102558-2729-Q7 NanoPore Reads Using DIAMOND BLASTx on Mox and MEGAN6 daa2rma on emu\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.bairdi NanoPore 6129-403-26 Quality Filtering Using NanoFilt on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.bairdi NanoPore 20102558-2729 Quality Filtering Using NanoFilt on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Assessment - BUSCO C.bairdi Genome v1.01 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 24, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Subsetting cbai_genome_v1.0 Assembly with faidx\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submissions - NanoPore C.bairdi 20102558-2729 and 6129_403_26\n\n\n\n\n\n\n\n2020\n\n\nSRA Submission\n\n\n\n\n\n\n\n\n\n\n\nSep 21, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Assessment - BUSCO C.bairdi Genome v1.0 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.bairdi NanoPore Quality Filtering Using NanoFilt on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - C.bairdi - cbai_v1.0 - Using All NanoPore Data With Flye on Mox\n\n\n\n\n\n\n\n2020\n\n\nGenome Assembly\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTaxonomic Assignments - C.bairdi NanoPore Reads Using DIAMOND BLASTx on Mox and MEGAN6 daa2rma on swoose\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Geoduck Normalizing Gene Primers 28s-v4 and EF1a-v1 Tests\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 16, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Geoduck Normalizing Gene Primer Checks\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Visualization of C.bairdi NanoPore Sequencing Using NanoPlot on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - NanoPore Fast5 Conversion to FastQ of C.bairdi 6129_403_26 on Mox with GPU Node\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 4, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - NanoPore Fast5 Conversion to FastQ of C.bairdi 20102558-2729 Run-02 on Mox with GPU Node\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 4, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - NanoPore Fast5 Conversion to FastQ of C.bairdi 20102558-2729 Run-01 on Mox with GPU Node\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 4, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submission - Supplemental Ronits C.gigas Diploid-Triploid Ctendidia gDNA for WGBS by ZymoResearch\n\n\n\n\n\n\n\n2020\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nSep 1, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Re-quant Ronits C.gigas Diploid-Triploid Ctenidia gDNA Submitted to ZymoResearch\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 1, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi v2.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nAug 31, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi v3.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nAug 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate Hematodinium v3.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate Hematodinium v2.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - C.bairdi Transcriptomes v2.1 and v3.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nAug 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - P.generosa RPL5 and TIF3s6b v2 and v3 Normalizing Gene Assessment\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 25, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submitted - C.gigas Diploid-Triploid pH Treatments Ctenidia to ZymoResearch for WGBS\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 24, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - P.generosa RPL5-v2-v3 and TIF3s6b-v2-v3 Primer Tests\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 24, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - C.gigas High-Low pH Triploid and Diploid Ctenidia\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 21, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - C.gigas High-Low pH Triploid Diploid from Maria Haws Lab\n\n\n\n\n\n\n\n2020\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nAug 20, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Ronits C.gigas Diploid and Triploid Ctenidia to ZymoResearch for WGBS\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 20, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Stats - C.bairdi Transcriptomes v2.1 and v3.1 Trinity Stats on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming-FastQC-MultiQC - Robertos C.gigas WGBS FastQ Data with fastp FastQC and MultiQC on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - Hematodinium Transcriptomes v1.6, v1.7, v2.1 and v3.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Stats - cbaiodinium Transcriptomes v2.1 and v3.1 Trinity Stats on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on Hematodinium v1.6 v1.7 v2.1 and v3.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Hematodinium Transcriptomes v1.6 v1.7 v2.1 v3.1 with DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - P.generosa APLP and TIF3s8-1 with cDNA\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQ Read Alignment and Quantification - P.generosa Water Metagenomic Libraries to MetaGeneMark Assembly with Hisat2 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPrimer Design and In-Silico Testing - Geoduck Reproduction Primers\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 30, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Testing P.generosa Reproduction-related Primers\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 29, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - P.generosa Metagenomics Data\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Testing P.generosa Reproduction-related Primers\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - C.gigas Diploid (Ronit) and Triploid (Nisbet)\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Geoduck Epigenetic Ocean Acidification RNAseq\n\n\n\n\n\n\n\n2020\n\n\nSRA Submission\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nENA Submission - Ostrea lurida draft genome Olurida_v081.fa\n\n\n\n\n\n\n\n2020\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Data Extractions Using MEGAN6\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Transcriptomes v2.1 and v3.1 Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v3.1\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v2.1\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequence Extractions - C.bairdi Transcriptomes v2.0 and v3.0 Excluding Alveolata with MEGAN6 on Swoose\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Transcriptomes v2.0 and v3.0 with DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 4, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Comparison - C.bairdi Transcriptomes Evaluations with DETONATE on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Comparison - C.bairdi Transcriptomes Compared with DETONATE on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 29, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi Transcriptome-v1.7 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 29, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Comparisons - C.bairdi BUSCO Scores\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - C.bairdi Transcriptome v1.7 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Transcriptome v1.7 Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v1.7\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi All Pooled Arthropoda-only RNAseq Data with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi Transcriptome-v3.0 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - P.trituberculatus (Japanese blue crab) NCBI SRA BioProject PRJNA597187 Data with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Library Assessment - Determine RNAseq Library Strandedness from P.trituberculatus SRA BioProject PRJNA597187\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 21, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTutorial - SRA Toolkit for Data Retrieval and Conversion to FastQ\n\n\n\n\n\n\n\n2020\n\n\nTutorials\n\n\n\n\n\n\n\n\n\n\n\nMay 21, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi Transcriptome-v1.6 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 20, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - C.bairdi Transcriptome v1.6 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - C.bairdi Transcriptome v3.0 from 20200518 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Transcriptome v1.6 Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v1.6\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Transcriptome v3.0 Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v3.0\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi All Arthropoda-specific RNAseq Data with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Arthropoda and Alveolata D26 Pool RNAseq FastQ Extractions\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi All Pooled RNAseq Data Without Taxonomic Filters with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi Transcriptome v2.0 from 20200502 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 13, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - C.bairdi Transcriptome v2.0 from 20200502 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Transcriptome v2.0 Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi v2.0 Transcriptome\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi All RNAseq Data Without Taxonomic Filters with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Abundance - C.bairdi Alignment-free with Salmon Using 2020-GW Data on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 29, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGO to GOslim - C.bairdi Enriched GO Terms from 20200422 DEGs\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQC - Laura Spencer’s QuantSeq Data\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGene Expression - C.bairdi Pairwise DEG Comparisons with 2019 RNAseq using Trinity-Salmon-EdgeR on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Reads Extractions - C.bairdi Taxonomic Reads Extractions with MEGAN6 on swoose\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Abundance - C.bairdi Alignment-free with Salmon on Mox for Grace\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - C.bairdi RNAseq Data\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimmingFastQCMultiQC—C.bairdi-RNAseq-FastQ-with-fastp-on-Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTaxonomic Assignments - C.bairdi RNAseq Using DIAMOND BLASTx on Mox and MEGAN6 Meganizer on swoose\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQC - C.bairdi Raw RNAseq from NWGSC\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Arthropoda and Alveolata Day and Treatment Taxonomic RNAseq FastQ Extractions\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.bairdi RNAseq from NWGSC\n\n\n\n\n\n\n\n2020\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi MEGAN6 Taxonomic-specific Trinity Assembly on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi MEGAN Trinity Assembly Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate Hematodinium MEGAN6 Taxonomic-specific Trinity Assembly on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransdecoder - Hematodinium MEGAN6 Taxonomic-Specific Reads Assembly from 20200330\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransdecoder - C.bairdi MEGAN6 Taxonomic-Specific Reads Assembly from 20200330\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi MEGAN Transcriptome\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Hematodinium MEGAN Trinity Assembly Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 31, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on Hematodinium MEGAN Transcriptome\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 31, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Hematodinium with MEGAN6 Taxonomy-specific Reads with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 30, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi with MEGAN6 Taxonomy-specific Reads with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 30, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Reads Extractions - C.bairdi Taxonomic Reads Extractions with MEGAN6 on swoose\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 30, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Using DIAMOND BLASTx on Mox and MEGAN6 Meganizer on swoose\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming/FastQC/MultiQC - C.bairdi RNAseq FastQ with fastp on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.bairdi RNAseq Data from Genewiz\n\n\n\n\n\n\n\n2020\n\n\nData Received\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - C.bairdi Hemocyte Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 11, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoPore Sequencing - C.bairdi gDNA 6129_403_26\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 11, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.bairdi RNA Check for Residual gDNA\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming/MultiQC - Methcompare Bisulfite FastQs with fastp on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 6, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - C.bairdi RNA from Hemolymph Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 6, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Create Canonical Olurida_v081 Genes FastA\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.bairdi RNA Check for Residual gDNA\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.bairdi Primer Tests on gDNA\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 25, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPrimer Design - C.bairdi Primers for Checking RNA for Residual gDNA\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 20, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - C.bairdi RNA from Samples 6212_132_9 6212_334_12 6212_485_26\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 11, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation & Quantification - C.bairdi RNA from Samples 6212_132_9 6212_334_12 6212_485_26\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 11, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation & Quantification - C.bairdi RNA from Sample 6129_403_26\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - Additional C.bairdi gDNA from Sample 6129_403_26\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation, Quantification, and Gel - C.bairdi gDNA Sample 6129_403_26\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on Hematodinium MEGAN Transcriptome\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi MEGAN Transcriptome\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGene Expression - Hematodinium MEGAN6 with Trinity and EdgeR\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGene Expression - C.bairdi MEGAN6 with Trinity and EdgeR\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Arthropoda and Alveolata Day and Treatment Taxonomic RNAseq FastQ Extractions\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 31, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - C.bairdi Hemocyte Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate Hematodinium MEGAN6 Taxonomic-specific Trinity Assembly on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi MEGAN6 Taxonomic-specific Trinity Assembly on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - C.bairdi Hemocyte Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - C.bairdi Hemocyte Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 24, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransdecoder - Hematodinium MEGAN6 Taxonomic-Specific Reads Assembly from 20200122\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransdecoder - C.bairdi MEGAN6 Taxonomic-Specific Reads Assembly from 20200122\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Hematodinium MEGAN Trinity Assembly Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi MEGAN Trinity Assembly Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - C.bairdi Hemocyte Pellets in RNAlater Troubleshooting\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quality Assessment - Agarose Gel for C.bairdi 20102558-2729 gDNA from 20200122\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Arthropoda and Alveolata Taxonomic RNAseq FastQ Extractions\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.bairdi 20102558-2729 EtOH-preserved Tissue via Three Variations Using Quick DNA-RNA MicroPrep Kit\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Hematodinium with MEGAN6 Taxonomy-specific Reads with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi with MEGAN6 Taxonomy-specific Reads with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - C.bairdi Hemolymph Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - C.bairdi Hemolymph Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Reads Extractions - C.bairdi Taxonomic Reads Extractions with MEGAN6 on swoose\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLab Maintenance - Cluster UPS Battery Replacement\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoPore Sequencing - C.bairdi gDNA Sample 20102558-2729\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quality Assessment - Agarose Gel and NanoDrop on C.bairdi gDNA\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - C.bairdi gDNA from EtOH Preserved Tissue\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoPore Sequencing - Initial NanoPore MinION Lambda Sequencing Test\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Using DIAMOND BLASTx on Mox and MEGAN6 Meganizer\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 3, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReagent Prep - RNA Pico Ladder Aliquoting and Testing\n\n\n\n\n\n\n\n2020\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nJan 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Trinity Assembly Trinotate on Mox\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Trinity Assembly BLASTx on Mox\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 24, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransdecoder - C.bairdi De Novo Transcriptome from 20191218 on Mox\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 20, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi Trimmed RNAseq Using Trinity on Mox\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming/FastQC/MultiQC - C.bairdi RNAseq FastQ with fastp on Mox\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Olurida_v081 UTR GFFs and Intergenic, Intron BED files\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 14, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Pacific Oyster Tissues from Hawaii (Maria Haws) from High and Low pCO2\n\n\n\n\n\n\n\n2019\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nDec 10, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Renaming, Splitting, and Feature Counts of Updated Pgenerosa_v074 GenSAS Merged GFF\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 3, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Crassostrea gigas and sikamea Mantle gDNA from Marinelli Shellfish Company\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 3, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - C.bairdi Hemolymph and Tissue in Ethanol from Pam Jensen\n\n\n\n\n\n\n\n2019\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nNov 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Geoduck hemolymph and hemocyte cDNA with vitellogenin primers\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - P.generosa DNased Hemolypmh and Hemocyte RNA from 20191125\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - Geoduck hemolymph and hemocyte samples\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Crassostrea gigas and sikamea Mantle gDNA from Marinellie Shellfish Company - No Multiplex\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Crassostrea gigas and sikamea Mantle gDNA from Marinelli Shellfish Company\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Rename Pgenerosa_v074 Bismark Coverage Files Scaffold Names\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 20, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Crassostrea gigas and sikamea Mantle gDNA from Marinelli Shellfish Company\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 19, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - Crassostrea gigas and Crassostrea sikamea Mantle Tissue from Marinelli Shellfish Company\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 19, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Additional Features Stats for Panopea-generosa-v1.0\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 6, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Rename Pgenerosa_v074 Files and Scaffolds\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 5, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Splitting BAM by Size for Upload to OSF\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 31, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Marinelli Shellfish Company C.gigas and C.sikamea Oysters\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 30, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Create Panopea-generosa-vv0.74.a4 Intron and Intergenic BED Files\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 30, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Feature Counts - Panopea-generosa-vv0.74.a4\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 29, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQC - C.bairdi RNAseq Day 12 26 Infected Uninfected\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nOct 24, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.bairdi RNAseq Day9-12-26 Infected-Uninfected\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nOct 24, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLab Maintenance - Cluster UPS Battery Replacement\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 22, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics Annotation - P.generosa Water Samples with MEGAN6\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 14, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.bairdi RNAseq Day9-12-26 Infected-Uninfected\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nOct 3, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v074 a4 Using GenSAS\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics Annotation - P.generosa Water Samples Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming/FastQC/MultiQC - P.generosa EPI FastQs with FASTP on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Hollie’s Juvenile OA BS-seq Data\n\n\n\n\n\n\n\n2019\n\n\nSRA Submissions\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Pgenerosa_v074.a3 Annotation Genome Repeats Compostion\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 5, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Pgenerosa_v074.a3 Annotation Genome Feature Sequence Lengths\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 4, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Panopea generosa Genome Feature Sequence Lengths\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Create a CpG GFF from Pgenerosa_v074 using EMBOSS fuzznuc on Swoose\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v074 MAKER on Mox with Stringtie Transcripts GFF\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 19, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs Pgenerosa_v070 with MUMmer Promer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs S.glomerata NCBI with MUMmer Promer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs M.yessoensis NCBI with MUMmer Promer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs H.sapiens NCBI with MUMmer Promer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs C.virginica NCBI with MUMmer Promer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs C.gigas NCBI with MUMmer Promer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs Pgenerosa_v070 with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs Pgenerosa_v074 with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs S.glomerata NCBI with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs M.yessoensis NCBI with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs C.gigas NCBI with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs H.sapiens NCBI with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs C.virginica NCBI with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeatMasker - Pgenerosa_v070 for Transposable Element ID on Roadrunner\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - FastA Splitting With faSplit\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Summary - P.generosa Transcriptome Assemblies Stats\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 29, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Compression - P.generosa Transcriptome Assemblies Using CD-Hit-est on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 29, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v074 Transcript Isoform ID with Stringtie on Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v070 Transcript Isoform ID with Stringtie on Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v074 Hisat2 Transcript Isoform Index\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v070 Hisat2 Transcript Isoform Index\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v070 and v074 Top 18 Scaffolds Feature Count Comparisons\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 22, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - O.lurida 20190709-v081 Transcript Isoform ID with Stringtie on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 16, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - O.lurida 20190709-v081 Hisat2 Transcript Isoform Index\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 16, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assessment - BUSCO Metazoa on Pgenerosa_v074 on Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assessment - BUSCO Metazoa on Pgenerosa_v70 on Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v071 Using GenSAS\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v074 Using GenSAS\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Olurida_v081 with MAKER and Tissue-specific Transcriptomes on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v074 MAKER on Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 1, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Juvenile Ambient OA EPI123 with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Juvenile Super Low OA EPI115 with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Larvae Day5 EPI99 with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Gonad with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Juvenile Ambient OA EPI124 with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Ctenidia with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Super Low OA EPI116 with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeatModeler - Pgenerosa_v074 for MAKER Annotation on Emu\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeatMasker - Pgenerosa_v074 for Transposable Element ID on Roadrunner\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.virginica Mantle MBD-BSseq from ZymoResearch\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - FastA Subsetting of Pgenerosa_v070.fa Using samtools faidx\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - O.lurida (v081) Transcript Isoform ID with Stringtie on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - O.lurida (v081) Hisat2 Transcript Isoforms Index\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Refining Anvio Binning\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 19, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Taxonomic Diversity and Sequencing Coverage with MEGAHIT BLASTx and Krona Plots\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 12, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Create Pgenerosa_v070 GFFs\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Tanner Crab Infected vs Uninfected RNAseq\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nMay 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - BLASTx of Individual Water Sample MEGAHIT Assemblies on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Decisions - C.bairdi RNAs for Library Pools\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQC - Additional C.gigas WGBS Sequencing Data from Genewiz Received 20190501\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 1, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Additional C.gigas Whole Genome Bisulfite Sequencing Data from Genewiz\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 1, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - C.bairdi Hemolymph Pellet in RNAlater\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 30, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - P.generosa 10x Genomics Sequencing Data\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 24, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - C.virginica MBD Sequencing Coverage\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics Annotation - P.generosa Water Samples Using BLASTn on Mox and KronaTools Visualization to Compare pH Treatments\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 16, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics Gene Prediction - P.generosa Water Samples Using MetaGeneMark on Mox to Compare pH Treatments\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 16, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC - WGBS Sequencing Data from Genewiz Received 20190408\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenome Assemblies - P.generosa Water Samples Trimmed HiSeqX Data Using Megahit on Mox to Compare pH Treatments\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - Crab Hemolypmh Using Quick-DNA-RNA Microprep Plus Kit\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nApr 11, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Larvae Day5 EPI99 with HiSeq and NovaSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Gonad HiSeq and NovaSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Juvenile Ambient OA EPI124 with HiSeq and NovaSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Juvenile Ambient OA EPI123 with HiSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Juvenile Super Low OA EPI116 with HiSeq and NovaSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Juvenile Super Low OA EPI115 with HiSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Ctenidia with HiSeq and NovaSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Whole Genome Bisulfite Sequencing Data from Genewiz Received\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - P.generosa Water Sample Assembly Comparisons with Quast\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Geoduck Water Sample Assembly Comparisons with MetaQuast\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 3, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Taxonomic Diversity Comparisons from Geoduck Water with Anvio on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 1, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenome Assemblies - P.generosa Water Samples Trimmed HiSeqX Data Using Megahit on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransposable Element Mapping - Crassostrea gigas Genome v9 Using RepeatMasker 4.07 on Roadrunner\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Lotterhos C.virginica Mantle MBD DNA to ZymoResearch for BSseq\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - SRA Submission Panopea generosa Day 5 Larvae RNAseq\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Taxonomic Diversity from Geoduck Water with BLASTn and Krona Plots\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Taxonomic Diversity from Geoduck Water with BLASTp and Krona plots\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment - DNA Quantification of C.virginica MBD Samples from 20190312\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 19, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Juvenile Day 5 with Trinotate on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Heart with Trinotate on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Ctenidia with Trinotate on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Gonad with Trinotate on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Heart with BLASTx on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Gonad with BLASTx on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Ctenidia with BLASTx on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Juvenile Day 5 with BLASTx on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Juvenile Day 5 with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Heart with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Gonad with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Ctenidia with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMethylation Analysis - C.virginica Gonad MBD with Varying Read Subsets with Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Create C.virginica Bisulfite Genome with Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 12, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment - Ethanol Precipitation of C.virginica MBD samples\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 12, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment - C.virginica Sheared Mantle DNA from 20190306\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 11, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment - C.virginica Sheared Mantle DNA from 20190306\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing & Bioanalyzer - Lotterhos C.virginica Mantle gDNA from 2018114\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 6, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Lotterhos C.virginica Mantle DNA from 20181114\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 5, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Data Migration and Drive Expansion on Gannet\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 4, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assessment - BUSCO Metazoa on P.generosa v071 on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v070 MAKER on Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - CpG OE Calculations on C.virginica Genes\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMethylation Analysis - O.lurida Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 22, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMethylation Analysis - P.generosa Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 22, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMethylation Analysis - C.virginica\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 22, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMethylation Analysis - C.gigas Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 22, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Create C.virginica Bisulfite Genome wit Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Create C.gigas Bisulfite Genome with Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Create Geoduck Bisulfite Genomes with Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Juvenile Day 5\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Heart\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Gonad\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Ctenidia\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v71 with MAKER on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequence Subsetting - Pgenerosa_v70 Genome Assembly with faidx\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 11, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Robertos C.gigas DNA for Whole Genome Bisulfite Sequencing (Genewiz)\n\n\n\n\n\n\n\n2019\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - C.virginica DNA and Tissues from Lotterhos Lab\n\n\n\n\n\n\n\n2019\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nJan 29, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - Ronit’s C.gigas Ploidy Ctenidia\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas Ploidy Experiment Ctenidia\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 17, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER BUSCO Metazoa Augustus Training on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Geoduck Genome with MAKER Submitted to Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER BUSCO (eukaryota_odb9) Augustus Training Submitted to Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER Functional Annotations on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER Proteins BLASTp on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER Proteins InterProScan5 on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER ID Mapping\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGene Prediction - HiSeqX Metagenomics from Geoduck Water Using MetaGeneMark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 3, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenome Assembly - P.generosa Water Sample Trimmed HiSeqX Data Using Megahit on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 2, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nVCF Splitting - C.virginica VCF Using BCFtools\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 2, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas Ploidy Experiment Ctenidia\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 20, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeat Library Construction - P.generosa RepeatModeler v1.0.11\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Relative mitochondrial abundance in C.gigas diploids and triploids subjected to acute heat stress via COX1\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBLASTx - Clupea pallasii (Pacific herring) liver and testes transcriptomes on Mox\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 12, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC and Trimming - Metagenomics (Geoduck) HiSeqX Reads from 20180809\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPrimer Design - Gigas COX1 using Primer3\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Geoduck gonad cDNA with vitellogenin primers\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 6, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Geoduck gonad RNA pool\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 6, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPrimer Design - Geoduck Vitellogenin using Primer3\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 29, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER on Mox\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 27, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Geoduck Transcritpome with TransDecoder\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 21, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Quantification - Ronit’s C.gigas DNased RNA from 20181115\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 20, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Ronit’s C.gigas DNased RNA from 20181115\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 20, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Ronit’s DNased C.gigas RNA with Elongation Factor Primers\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 15, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - Ronit’s C.gigas Ctenidia RNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 15, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - Lotterhos C.virginica Mantle DNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Install NCBI nr nt BLAST Database on Mox\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMy New Notebook\n\n\n\n\n\n\n\n2018\n\n\n\n\n\n\n\n\n\n\n\n\n\n\nNov 8, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeat Library Construction - O.lurida RepeatModeler v1.0.11\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 22, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - Ronit’s C.gigas ploidy/dessication/heat stress cDNA (1:5 dilution)\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 18, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Crassostrea virginica (Eastern oyster) tissue from Lotterhos Lab (Northeastern University)\n\n\n\n\n\n\n\n2018\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nOct 17, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Ronit’s C.gigas DNased ctenidia RNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 17, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Ronit’s DNAsed C.gigas Ploidy/Dessication RNA with elongation factor primers\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 17, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR – C.gigas primer and gDNA tests with 18s and EF1 primers\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Ronit’s DNAsed C.gigas Ploidy/Dessication RNA with 18s primers\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Ronit’s C.gigas Ploiyd/Dessication Ctenidia RNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Quantification - Ronit’s C.gigas Ploidy/Dessication RNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Chionoecetes bairdi RNAseq & FastQC Analysis\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 15, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nVCF Splitting with bcftools\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 15, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Tanner Crab Hemolymph Pellet in RNAlater using TriReagent\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Ronit’s C.gigas diploid/triploid dessication/heat shock ctenidia tissues\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 3, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Olymia oyster Whole Genome BS-seq Data\n\n\n\n\n\n\n\n2018\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 3, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nInstallation - Microsoft Machine Learning Server (Microsoft R Open) on Emu/Roadrunner R Studio Server\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nUbuntu – Fix “No Video Signal” Issue on Emu\n\n\n\n\n\n\n\n2018\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Alignment & Bedgraph – Olympia oyster transcriptome with Olurida_v080 genome assembly\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 26, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBedgraph – Olympia oyster transcriptome with Olurida_v081 genome assembly\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 26, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Alignment – Olympia oyster RNAseq reads aligned to genome with HISAT2\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 25, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBedgraph - Olympia oyster transcriptome (FAIL)\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 24, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Alignment - Olympia oyster Trinity transcriptome aligned to genome with Bowtie2\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Olympia oyster RNAseq Data with Trinity\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Analysis – Olympia Oyster Whole Genome BSseq Bismark Pipeline MethylKit Comparison\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Lyophilized Tanner Crab Hemolymph in RNAlater\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Analysis - Olympia Oyster Whole Genome BSseq Bismark Pipeline Comparison\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 13, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Sea Lice DNA from 20180523\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 12, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC - C.virginica Oil Spill MBDseq Concatenated Sequences\n\n\n\n\n\n\n\n2018\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing Data Analysis - C.virginica Oil Spill MBDseq Concatenation & FastQC\n\n\n\n\n\n\n\n2018\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 10, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Analysis - Olympia oyster BSseq MethylKit Analysis\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 4, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck RNAseq data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 4, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC/MultiQC/TrimGalore/MultiQC/FastQC/MultiQC - O.lurida WGBSseq for Methylation Analysis\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 30, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransposable Element Mapping – Crassostrea virginica Genome, Cvirginica_v300, using RepeatMasker 4.07\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 28, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Stats – Geoduck Hi-C Final Assembly Comparison\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 23, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Analysis - Bismark Pipeline on All Olympia oyster BSseq Datasets\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Geoduck Metagenome HiSeqX Data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation & Quantificaiton - Tanner Crab Hemolymph\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation – Olympia oyster genome annotation results #02\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 8, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Olympia oyster genome annotation results #01\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 8, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation – Olympia oyster genome complete - brief note\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Olympia oyster genome using WQ-MAKER Instance on Jetstream\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - Tanner Crab RNA Isolated with RNeasy Plus Mini Kit\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 1, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Cleanup - Tanner Crab RNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Tanner Crab Hemolymph Using RNeasy Plus Mini Kit\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMox – Over quota: Olympia oyster genome annotation progress (using Maker 2.31.10)\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 30, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Cleanup - Tanner Crab RNA Pools\n\n\n\n\n\n\n\n2018\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJul 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMox - Olympia oyster genome annotation progress (using Maker 2.31.10)\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMox - Password-less SSH!\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nUbuntu - Fix “No Video Signal” Issue on Emu/Roadrunner\n\n\n\n\n\n\n\n2018\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJul 5, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransposable Element Mapping – Olympia Oyster Genome Assembly, Olurida_v081, using RepeatMasker 4.07\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 3, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Construction - Geoduck Water Filter Metagenome with Nextera DNA Flex Kit (Illumina)\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 6, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBS-seq Mapping – Olympia oyster bisulfite sequencing: Bismark Continued\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 31, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransposable Element Mapping – Crassostrea virginica NCBI Genome Assembly using RepeatMasker 4.07\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 29, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Mapping – Olympia oyster 2bRAD Data with Bowtie2 (on Mox)\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 26, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransposable Element Mapping - Olympia Oyster Genome Assembly using RepeatMasker 4.07\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Received - Sea lice DNA from Cris Gallardo-Escarate at Universidad de Concepción\n\n\n\n\n\n\n\n2018\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Installation – RepeatMasker v4.0.7 on Emu/Roadrunner Continued\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Installation - RepeatMasker v4.0.7 on Emu/Roadrunner\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 22, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC – TrimGalore! RRBS Geoduck BS-seq FASTQ data (directional)\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC - RRBS Geoduck BS-seq FASTQ data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC – TrimGalore! RRBS Geoduck BS-seq FASTQ data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Illumina NovaSeq Geoduck Genome Sequencing\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly - Geoduck Hi-C Assembly Subsetting\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 12, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Mapping - Mapping Illumina Data to Geoduck Genome Assemblies with Bowtie2\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBS-seq Mapping - Olympia oyster bisulfite sequencing: TrimGalore > FastQC > Bismark\n\n\n\n\n\n\n\n2018\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly & Stats - SparseAssembler (k95) on Geoduck Sequence Data > Quast for Stats\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 1, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Stats - Geoduck Genome Assembly Comparisons w/Quast - SparseAssembler, SuperNova, Hi-C\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 30, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Stats - Geoduck Hi-C Assembly Comparison\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 30, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - Metagenomics Water Filters\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTotal Alkalinity Calculations - Yaamini’s Ocean Chemistry Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 24, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly – SparseAssembler (k 111) on Geoduck Sequence Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 23, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly – SparseAssembler (k 131) on Geoduck Sequence Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 22, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Geoduck Phase Genomics Hi-C Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nApr 21, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nKmer Estimation – Kmergenie (k 301) on Geoduck Sequence Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 21, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nKmer Estimation – Kmergenie Tweaks on Geoduck Sequence Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nKmer Estimation - Kmergenie on Geoduck Sequence Data (default settings)\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Stats - Quast Stats for Geoduck SparseAssembler Job from 20180405\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - SRA Submission LSU C.virginica Oil Spill MBD BS-seq Data\n\n\n\n\n\n\n\n2018\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 12, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC - Trim 10bp 5’/3’ ends C.virginica MBD BS-seq FASTQ data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - Metagenomics Water Filters\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC - 2bp 3’ end Read 1s Trim C.virginica MBD BS-seq FASTQ data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC - 14bp Trim C.virginica MBD BS-seq FASTQ data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC - Auto-trim C.virginica MBD BS-seq FASTQ data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC/MultiQC - C. virginica MBD BS-seq Data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - SparseAssembler Geoduck Genomic Data, kmer=101\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 5, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGunzip - BGI HiSeq Geoduck Genome Sequencing Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 5, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGunzip - Trimmed Illumina Geoduck HiSeq Genome Sequencing Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 4, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification – Geoduck larvae metagenome filter rinses\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 3, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrations - Yaamini’s Seawater Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 3, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrations - Yaamini’s Seawater Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 2, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore!/FastQC/MultiQC - Illumina HiSeq Genome Sequencing Data Continued\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 1, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Crassostrea virginica MBD BS-seq from ZymoResearch\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 29, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC/MultiQC – Illumina HiSeq Genome Sequencing Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 28, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore!/FastQC/MultiQC - Illumina HiSeq Genome Sequencing Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 28, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC/MultiQC - BGI Geoduck Genome Sequencing Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 27, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrations - Hollie’s Seawater Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 27, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly - Geoduck NovaSeq using SparseAssembler kmer = 101\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 27, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly - Geoduck NovaSeq using SparseAssembler (TL;DR - it worked!)\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 22, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrations - Hollie’s Seawater Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 20, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - Geoduck larvae metagenome filter rinses\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 20, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrations - Hollie’s Seawater Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrations - Hollie’s Seawater Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Geoduck larvae metagenome filter rinses\n\n\n\n\n\n\n\n2018\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nMar 13, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly - Geoduck NovaSeq using SparseAssembler (failed)\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 8, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nProgress Report - Titrator\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 6, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nUbuntu Installation - Convert Apple Xserve “bigfish” to Ubuntu\n\n\n\n\n\n\n\n2018\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHardware Upgrades - USB 3.0 PCI Card and 1TB SSD in Woodpecker\n\n\n\n\n\n\n\n2018\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Triploid Crassostrea gigas from Nisbet Oyster Company\n\n\n\n\n\n\n\n2018\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNovaSeq Assembly - The Struggle is Real - Real Annoying!\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 21, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly - Geoduck Illumina NovaSeq SOAPdenovo2 on Mox (FAIL)\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation & DNA Quantification - C. virginica MBD DNA from Yaamini\n\n\n\n\n\n\n\n2018\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNovaSeq Assembly - Trimmed Geoduck NovaSeq with Meraculous\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 5, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrator Setup - Functional Methods & Data Exports\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 1, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAdapter Trimming and FASTQC - Illumina Geoduck Novaseq Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 25, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Install - 10x Genomics Supernova on Mox (Hyak)\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - C. virginica gDNA, MBD, and MspI to Qiagen\n\n\n\n\n\n\n\n2018\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nJan 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Comparisons – Oly Assemblies Using Quast\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - MspI-digested Crassostrea virginica gDNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPhenol:Chloroform Extractions and EtOH Precipitations - MspI Digestions of C.virginica DNA from Earlier Today\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digestion - MspI on Crassotrea virginica gDNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - C.virginica MBD-enriched DNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 10, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment – Crassostrea virginica Sheared DNA Day 3\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 10, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment – Crassostrea virginica Sheared DNA Day 2\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment - Crassostrea virginica Sheared DNA Day 1\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTissue Sampling - Crassostrea virginica Tissues for Archiving\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 13, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Install - MSMTP For Email Notices of Bash Job Completion on Emu (Ubuntu)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 13, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Pulverized Geoduck Tissues to Illumina for More 10x Genomics Sequencing\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 12, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Sonication & Bioanalzyer - C. virginica gDNA for MeDIP\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - Crassostrea virginica Mantle gDNA\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly – Olympia Oyster Illumina & PacBio Using PB Jelly w/BGI Scaffold Assembly\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTroubleshooting – PB Jelly Install on Emu Continued\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Geoduck Tissues to Illumina for More 10x Genomics Sequencing\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nNov 20, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTroubleshooting - PB Jelly Install on Emu\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 20, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - C. virginica Gonad gDNA\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Comparison - Oly Assemblies Using Quast\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia Oyster Illumina & PacBio Using PB Jelly w/BGI Scaffold Assembly\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia Oyster Illumina & PacBio Using PB Jelly w/Platanus Assembly\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Tanner Crab Hemolymph in RNA Later from Pam Jensen\n\n\n\n\n\n\n\n2017\n\n\nSamples Received\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation & Quantification - Tanner crab hemolymph\n\n\n\n\n\n\n\n2017\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nNov 7, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Installation - ALPACA on Roadrunner\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 31, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Crash - Olympia oyster genome assembly with Masurca on Mox\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 31, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Installation - PB Jelly Suite and Blasr on Emu\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 30, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster Illumina & PacBio Reads Using Redundans\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 24, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Comparison - Oly PacBio Canu: Sam vs. Sean with Quast\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 23, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFAIL - Missing Data on Owl!\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 23, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster Illumina & PacBio reads using MaSuRCA\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 19, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Installation - MaSuRCA v3.2.3 Assembler on Mox (Hyak)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 19, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFail - Directory Contents Deleted!\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 18, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster PacBio Canu v1.6\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 18, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Convert Oly PacBio H5 to FASTQ\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster Redundans/Canu vs. Redundans/Racon\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 6, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia Oyster Redundans with Illumina + PacBio\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - minimap/miniasm/racon Overview\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 4, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Comparisons - Olympia oyster genome assemblies\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 3, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - C.virginica gonad tissue from Katie Lotterhos\n\n\n\n\n\n\n\n2017\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nOct 3, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - October 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster PacBio minimap/miniasm/racon\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster PacBio minimap/miniasm/racon\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster PacBio minimap/miniasm/racon\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 7, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Geoduck Ctenidia to Illumina for 10x Genomics Sequencing\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nAug 28, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nProject Progress - Olympia Oyster Genome Assemblies by Sean Bennett\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 28, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Illumina Geoduck HiSeq & MiSeq Data\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 8, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - August 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nAug 3, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Geoduck Genome Sequencing by Illumina\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Olympia oyster gonad RNA to Katherine Silliman @ Univ. of Chicago\n\n\n\n\n\n\n\n2017\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nJul 20, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Olympia oyster gonad tissue in paraffin histology blocks\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 19, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Olympia oyster gonad tissue in paraffin histology blocks\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - SRA Submission Olympia Oyster UW PacBio Data from 20170323\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management – Tarball of Olympia oyster UW PacBio Data from 20170323\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - OLYMPIA OYSTER UW PACBIO DATA (FROM 20170323) TO NIGHTINGALES\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 5, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Annotation - Olympia oyster histology blocks (from Laura Spencer)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 5, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nManuscript Re-submission - Oly Stress Response to PeerJ for Review\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 3, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Olympia oyster UW PacBio Data from 20170323\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 3, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGitHub Curation\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 22, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - June 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Quantification - Acropora cervicornis (Staghorn coral) DNA from Javier Casariego (FIU)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 11, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Acropora cervicornis (Staghorn coral) DNA from Javier Casariego (FIU)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 11, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Concentration - Acropora cervicornis (Staghorn coral) DNA from Javier Casariego (FIU)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 10, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Acropora cervicornis (Staghorn coral) DNA from Javier Casariego (FIU)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 10, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Olympia oyster Histology Blocks and Slides (for Laura Spencer)\n\n\n\n\n\n\n\n2017\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nMay 9, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Acropora cervicornis (Staghorn coral) DNA from Javier Casariego (FIU)\n\n\n\n\n\n\n\n2017\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nMay 9, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - May 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nMay 2, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nManuscript Writing - Submitted!\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nApr 19, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nManuscript - Oly GBS 14 Day Plan\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nApr 18, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Olympia oyster PacBio Data\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 23, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management – SRA Submission Oly GBS Batch Submission\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 21, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Owl Partially Restored\n\n\n\n\n\n\n\n2017\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nMar 21, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - SRA Submission Oly GBS Batch Submission Fail\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 20, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTroubleshooting - Synology NAS (Owl) Down After Update\n\n\n\n\n\n\n\n2017\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing – Oly BGI GBS Reproducibility; fail?\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 17, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing – Oly BGI GBS Reproducibility Fail (but, less so than last time)…\n\n\n\n\n\n\n\n2017\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Oly BGI GBS Reproducibility Fail\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 7, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFASTQC - Oly BGI GBS Raw Illumina Data Demultiplexed\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 7, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFASTQC - Oly BGI GBS Raw Illumina Data\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - March 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nManuscript Writing - More “Nuances” Using Authorea\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Jay’s Coral RADseq and Hollie’s Geoduck Epi-RADseq\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 27, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - SRA Submission of Ostrea lurida GBS FASTQ Files\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\nSRA Submissions\n\n\n\n\n\n\n\n\n\n\n\nFeb 8, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - February 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nFeb 2, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nManuscript Writing - The “Nuances” of Using Authorea\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission – Geoduck gDNA for Illumina Pilot Sequencing Project\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nJan 5, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck gDNA for Illumina-initiated Sequencing Project\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 5, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Replacement of Corrupt BGI Oly Genome FASTQ Files\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 4, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - January 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nJan 3, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Geoduck RRBS Data Integrity Verification\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 30, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Geoduck RRBS Sequencing Data\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 29, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Geoduck Tissue & gDNA for Illumina Pilot Sequencing Project\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 21, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck gDNA for Potential Illumina-initiated Sequencing Project\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 21, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Geoduck Reduced Representation Bisulfite Pooled Libraries\n\n\n\n\n\n\n\n2016\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 20, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Ostrea lurida gDNA for PacBio Sequencing\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 19, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Integrity Check of Final BGI Olympia Oyster & Geoduck Data\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 15, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Ostrea lurida DNA for PacBio Sequencing\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Managment - Trim Output Cells from Jupyter Notebook\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Download Final BGI Genome & Assembly Files\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - December 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Continued O.lurida Fst Analysis from GBS Data\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - An Excercise in Futility\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Initial O.lurida Fst Determination from GBS Data\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 17, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Tracking O.lurida FASTQ File Corruption\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 17, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputer Management - Additional Configurations for Reformatted Xserves\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nNov 16, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Modify Eagle/Owl Cloud Sync Account\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nNov 7, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Retrieve data from Amazon EC2 Instance\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 4, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - November 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nNov 2, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management – Geoduck Small Insert Library Genome Assembly from BGI\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 25, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - October 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nOct 3, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received – Jay’s Coral epiRADseq - Not Demultiplexed\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 19, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nOyster Sampling - Olympia Oyster OA Populations at Manchester\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 12, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Jay’s Coral epiRADseq\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 8, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - September 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nSep 6, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Synology Cloud Sync to UW Google Drive\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nAug 29, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nServer HDD Failure – Owl\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nAug 22, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nManuscript Submission - Oly Stress Response to PeerJ for Review\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 18, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - fastStructure Population Analysis of Oly GBS PyRAD Output\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nAug 16, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Amazon EC2 Cost “Analysis”\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 1, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - August 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nAug 1, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - PyRad Analysis of Olympia Oyster GBS Data\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nJul 27, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Not Enough Power!\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 18, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Amazon EC2 Instance Out of Space?\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nJul 17, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - A Very Quick “Guide” to Amazon EC2 Continued\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDissection - Frozen Geoduck & Pacific Oyster\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFilter Replacement - Xserve Server Rack Enclosure\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - The Very Quick “Guide” to Amazon Web Services Cloud Computing Instances (EC2)\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 13, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Quantification - Coral DNA from Jose M. Eirin-Lopez (Florida International University)\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 7, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - July 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nJul 1, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Coral DNA from Jose M. Eirin-Lopez (Florida International University)\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Coral DNA from Jose M. Eirin-Lopez (Florida International University)\n\n\n\n\n\n\n\n2016\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nJun 15, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDocker - VirtualBox Defaults on OS X\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJun 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDocker - One liner to create Docker container\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 9, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRAM Upgrade - Roadrunner (Apple Xserve) to 48GB RAM\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJun 9, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - June 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nJun 6, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDocker - Improving Roberts Lab Reproducibility\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputer Setup - Cluster Node003 Conversion\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nMay 31, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Oly GBS Data Using Stacks 1.37\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Olympia Oyster Small Insert Library Genome Assembly from BGI\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGBS Frustrations\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 3, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - May 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nMay 2, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Release - Transcriptomic Profiles of Adult Female & Male Gonads in Panopea generosa (Pacific geoduck)\n\n\n\n\n\n\n\n2016\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\nSRA Submissions\n\n\n\n\n\n\n\n\n\n\n\nApr 28, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - O.lurida Raw BGI GBS FASTQ Data\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nApr 27, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Speed Benchmark Comparisons Between Local, External, & Server Files\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 27, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Subset Olympia Oyster GBS Data from BGI as Single Population Using PyRAD\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nApr 18, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Concatenate FASTQ files from Oly MBDseq Project\n\n\n\n\n\n\n\n2016\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nApr 11, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Oly GBS Data from BGI Using Stacks\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nApr 6, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Install - samtools-0.1.19 and stacks-1.37\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nApr 6, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission – Genome sequencing of the Olympia oyster (Ostrea lurida)\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\nSRA Submissions\n\n\n\n\n\n\n\n\n\n\n\nMar 28, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission – Genome sequencing of the Pacific geoduck (Panopea generosa)\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\nSRA Submissions\n\n\n\n\n\n\n\n\n\n\n\nMar 28, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Transcriptomic Profiles of Adult Female & Male Gonads in Panopea generosa (Pacific geoduck).\n\n\n\n\n\n\n\n2016\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nMar 24, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Individual Transcriptomic Profiles of C.gigas Before & After Heat Shock\n\n\n\n\n\n\n\n2016\n\n\nSRA Submissions\n\n\n\n\n\n\n\n\n\n\n\nMar 23, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - SRA Submission Overview\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 23, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - O. lurida genotype-by-sequencing (GBS) data from BGI\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Initial Geoduck Genome Assembly from BGI\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Initial Olympia oyster Genome Assembly from BGI\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - O.lurida 2bRAD Dec2015 Undetermined FASTQ files\n\n\n\n\n\n\n\n2016\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nMar 8, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - High-throughput Sequencing Data\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 4, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Ostrea lurida MBD-enriched BS-seq\n\n\n\n\n\n\n\n2016\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nFeb 3, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Ostrea lurida genome sequencing files from BGI\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 27, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Panopea generosa genome sequencing files from BGI\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 27, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Identification of duplicate files on Eagle\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Bisulfite-treated Illumina Sequencing from Genewiz\n\n\n\n\n\n\n\n2016\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Oly 2bRAD Illumina Sequencing from Genewiz\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 31, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - BS-seq Library Pool to Genewiz\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nIllumina Methylation Library Quantification - BS-seq Oly/C.gigas Libraries\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nIllumina Methylation Library Construction - Oly/C.gigas Bisulfite-treated DNA\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 21, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - Bisulfite-treated Oly/C.gigas DNA\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReagent Prep - RNA Pico 6000 Ladder\n\n\n\n\n\n\n\n2015\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisulfite Treatment - Oly Reciprocal Transplant DNA & C.gigas Lotterhos DNA for BS-seq\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Oly gDNA for BS-seq Libraries, Take Two\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Oly gDNA for BS-seq Libraries\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Oly gDNA for BS-seq\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - 2bRAD Libraries for Genewiz\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 10, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - C.gigas Tissue & DNA from Katie Lotterhos\n\n\n\n\n\n\n\n2015\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nDec 10, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Olympia oyster MBD-enriched DNA to ZymoResearch\n\n\n\n\n\n\n\n2015\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\nOlympia oyster reciprocal transplant\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 8, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Additional Olympia Oyster gDNA for Genome Sequencing @ BGI\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Additional Geoduck gDNA for Genome Sequencing @ BGI\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Storage - Synology DX513\n\n\n\n\n\n\n\n2015\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Oly Oyster Bay Tissues for GBS\n\n\n\n\n\n\n\n2015\n\n\nGenotype-by-sequencing at BGI\n\n\nOlympia oyster reciprocal transplant\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Oly Tissue & DNA from Katherine Silliman\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quality Assessment - Geoduck & Olympia Oyster gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Olympia Oyster Outer Mantle gDNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck Ctenidia gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPhenol-Chloroform DNA Cleanup - Geoduck gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPhenol-Chloroform DNA Cleanup - Olympia Oyster gDNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck Adductor Muscle gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Olympia Oyster Outer Mantle gDNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - MBD-enriched Olympia oyster DNA\n\n\n\n\n\n\n\n2015\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 23, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation - Olympia oyster MBD\n\n\n\n\n\n\n\n2015\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment - Sonicated Olympia Oyster gDNA\n\n\n\n\n\n\n\n2015\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 21, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Sonication - Oly gDNA for MBD\n\n\n\n\n\n\n\n2015\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\n\n\n\n\n\n\n\n\n\nNov 19, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR – Oly RAD-Seq Library Quantification\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Oly RAD-Seq Library Quantification\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGel Extraction - Oly RAD-Seq Prep Scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quality Assessment - Geoduck, Oly & Oly 2SN\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR – Oly RAD-seq Prep Scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR – Oly RAD-seq Test-scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 12, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification & Quality Assessment - Oly 2SN gDNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 4, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification & Quality Assessment - Geoduck & Oly gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 4, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolations – Oly Fidalgo 2SN Ctenidia\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 3, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nOyster Sampling - Oly Fidalgo 2SN, 2HL, 2NF Reciprocal Transplants Final Samplings\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 30, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation – Geoduck & Olympia Oyster\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 29, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAdaptor Ligation – Oly AlfI-Digested gDNA for RAD-seq\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 29, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digest – Oly gDNA for RAD-seq w/AlfI\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 28, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTroubleshooting - Oly RAD-seq\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 23, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolations - Fidalgo 2SN Reciprocal Transplants Final Samplings\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 21, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Oly RAD-seq Prep Scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Oly RAD-seq Test-scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 12, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAdaptor Ligation – Oly AlfI-Digested gDNA for RAD-seq\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digest – Oly gDNA for RAD-seq w/AlfI\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Additional Geoduck gDNA for Genome Sequencing @ BGI\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Additional Olympia Oyster gDNA for Genome Sequencing @ BGI\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nOct 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Geoduck & Olympia Oyster gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Pooled geoduck gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Geoduck Adductor Muscle\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck & Olympia Oyster\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Oly RAD-seq Test-scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Oly RAD-seq Test-scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nOct 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAdaptor Ligation - Oly AlfI-Digested gDNA for RAD-seq\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nOct 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - October 2015\n\n\n\n\n\n\n\n2015\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nOct 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digest - Oly gDNA for RAD-seq w/AlfI\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nSep 30, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Olympia Oyster gDNA for Genome Sequencing @ BGI\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nSep 21, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Geoduck gDNA for Genome Sequencing @ BGI\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 21, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nUninterruptible Power Supplies (UPS)\n\n\n\n\n\n\n\n2015\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Geoduck & Olympia oyster gDNA Integrity Check\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation - Geoduck & Olympia oyster gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Olympia oyster Whole Body gDNA Integrity Check\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Olympia oyster whole body\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nSep 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel – Geoduck gDNA Integrity Check\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation – Geoduck Adductor Muscle & Foot\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Geoduck & Olympia Oyster gDNA Integrity Check\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation – Olympia oyster adductor musle & mantle\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation – Geoduck Adductor Muscle & Foot\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Geoduck & Olympia Oyster gDNA Integrity Check\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation - Olympia oyster adductor musle & mantle\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation - Geoduck Adductor Muscle & Foot\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Geoduck & Olympia Oyster gDNA Integrity Check\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation - Olympia oyster adductor musle & mantle\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nServer Email Notifications Fix - Eagle\n\n\n\n\n\n\n\n2015\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nSep 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation - Geoduck Adductor Muscle & Foot\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 28, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRAD-Seq Library Prep Reagents\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nAug 19, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSsoFast EvaGreen Supermix Aliquots\n\n\n\n\n\n\n\n2015\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nAug 11, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription – O.lurida DNased RNA 1hr post-mechanical stress\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Jake’s O.lurida ctenidia 1hr post-mechanical stress DNased RNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Quantification - O.lurida 1hr post-mechanical heat stress DNased RNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - August 2015\n\n\n\n\n\n\n\n2015\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nAug 3, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission – Olympia oyster PCRs Sanger Sequencing\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nServer HDD Failure - Owl\n\n\n\n\n\n\n\n2015\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - O.lurida Ctenidia 1hr Post-Mechanical Stress RNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nJul 27, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation – O.lurida Ctenidia 1hr Post-Mechanical Stress\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nJul 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - O.lurida Ctenidia 1hr Post-Mechanical Stress\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSsoFast EvaGreen Supermix Aliquots\n\n\n\n\n\n\n\n2015\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAutomatic Notebook Backups - wget Script & Synology Task Scheduler\n\n\n\n\n\n\n\n2015\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Sea Pen luciferase\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGOALS - July 2015\n\n\n\n\n\n\n\n2015\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nOpticon2 Calibration\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Data Receipt - Geoduck Gonad RNA 100bp PE Illumina\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission – Olympia oyster PCRs Sanger Sequencing\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - O.lurida DNased RNA Controls and 1hr Heat Shock\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission – Olympia oyster & Sea Pen PCRs Sanger Sequencing\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGel Purification - Olympia Oyster and Sea Pen PCRs\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Olympia oyster PCRs Sanger Sequencing\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 12, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Geoduck Gonad for RNA-seq\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - June 2015\n\n\n\n\n\n\n\n2015\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - Geoduck Gonad RNA Quality Assessment\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Subset of Jake’s O.lurida DNased RNA\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioinformatics – Trimmomatic/FASTQC on C.gigas Larvae OA NGS Data\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Re-run Jake’s O.lurida DNased RNA Samples NC1, SC1, SC2, SC4 from 20150514\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 21, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR – Jake’s O.lurida ctenidia DNased RNA (1hr Heat Shock Samples)\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR – Jake’s O.lurida ctenidia DNased RNA (Control Samples)\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Jake’s O.lurida Ctenidia RNA (1hr Heat Shock) from 20150506\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Jake’s O.lurida Ctenidia RNA (Controls) from 20150507\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nISO Creation - OpticonMonitor3 Disc Cloning\n\n\n\n\n\n\n\n2015\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Jake O.lurida ctenidia RNA (Heat Shock Samples) from 20150506\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 12, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Jake O.lurida ctenidia RNA (Control Samples) From 20150507\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 12, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation – Geoduck Gonad in Paraffin Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nMay 7, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation – Jake’s O. lurida Ctenidia Control from 20150422\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 7, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioinformatics - Trimmomatic/FASTQC on C.gigas Larvae OA NGS Data\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Jake’s O. lurida Ctenidia 1hr Heat Stress from 20150422\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation – Geoduck Gonad in Paraffin Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nMay 5, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBLAST - C.gigas Larvae OA Illumina Data Against GenBank nt DB\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 4, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - May 2015\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\nGoals\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nMay 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBLASTN - C.gigas OA Larvae to C.gigas Ensembl 1.24 BLAST DB\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 29, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation – Geoduck Gonad in Paraffin Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer Data - Geoduck RNA from Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nApr 24, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation – Geoduck Gonad in Paraffin Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nApr 23, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer Submission - Geoduck Gonad RNA from Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nApr 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nQuality Trimming - C.gigas Larvae OA BS-Seq Data\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nQuality Trimming - LSU C.virginica Oil Spill MBD BS-Seq Data\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequence Data Analysis - LSU C.virginica Oil Spill MBD BS-Seq Data\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequence Data Analysis - C.gigas Larvae OA BS-Seq Data\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequence Data - C.gigas OA Larvae BS-Seq Demultiplexed\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequence Data - LSU C.virginica Oil Spill MBD BS-Seq Demultiplexed\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Geoduck Gonad in Paraffin Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing Data - C.gigas Larvae OA\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 19, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEpinext Adaptor 1 Counts - LSU C.virginica Oil Spill Samples\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTruSeq Adaptor Counts – LSU C.virginica Oil Spill Sequences\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTruSeq Adaptor Identification Method Comparison - LSU C.virginica Oil Spill Sequences\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Claire’s C.gigas Sheared DNA\n\n\n\n\n\n\n\n2015\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nMar 3, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Quality Assessment - C.gigas OA larvae Illumina libraries\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBS-seq Library Prep - C.gigas Larvae OA 1000ppm\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 27, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - C.gigas Larvae 1000ppm\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 26, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Claire’s Sheared C.gigas Mantle Heat Shock Samples\n\n\n\n\n\n\n\n2015\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nFeb 26, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - C.gigas Sheared DNA from 20140108\n\n\n\n\n\n\n\n2015\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nFeb 20, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Prep - Quantification of C.gigas larvae OA 1000ppm library\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing Data - LSU C.virginica MBD BS-Seq\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisulfite NGS Library Prep - Bisulfite Conversion & Illumina Library Construction of C.gigas larvae DNA\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisuflite NGS Library Prep – C.gigas larvae OA bisulfite library quantification\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 28, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisuflite NGS Library Prep - C.gigas larvae OA bisulfite DNA (continued from yesterday)\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 27, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisuflite NGS Library Prep - C.gigas larvae OA bisulfite DNA\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 26, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Cleanup - LSU C.virginica MBD BS Library\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Bisulfite Conversion - C.gigas larvae OA Sheared DNA\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSpeedVac - C.gigas larvae OA DNA\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas larvae from 2011 NOAA OA Experiment\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisulfite NGS Library - LSU C.virginica Oil Spill MBD Bisulfite DNA Sequencing Submission\n\n\n\n\n\n\n\n2014\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisulfite NGS Library Prep - LSU C.virginica Oil Spill MBD Bisulfite DNA and Emma’s C.gigas Larvae OA Bisulfite DNA (continued from yesterday)\n\n\n\n\n\n\n\n2014\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisulfite NGS Library Prep - LSU C.virginica Oil Spill Bisulfite DNA and Emma’s C.gigas Larvae OA Bisulfite DNA\n\n\n\n\n\n\n\n2014\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisulfite Conversion - LSU C.virginica Oil Spill MBD DNA and Emma’s C.gigas Larvae OA DNA\n\n\n\n\n\n\n\n2014\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 16, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Claire’s C.gigas Female Gonad for Illumina Bisulfite Sequencing\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEtOH Precipitation - LSU C.virginica Oil Spill MBD Continued (from 20141126)\n\n\n\n\n\n\n\n2014\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 2, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMethylated DNA Enrichment (MBD) - LSU C.virginica Oil Spill gDNA\n\n\n\n\n\n\n\n2014\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 26, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing - LSU C.virginica Oil Spill gDNA\n\n\n\n\n\n\n\n2014\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGel - Sheared gDNA\n\n\n\n\n\n\n\n2014\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Seq - C.gigas Total RNA from Claire’s Pre/Post Heat Shock\n\n\n\n\n\n\n\n2014\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas Larvae from Emma OA Experiments\n\n\n\n\n\n\n\n2014\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 21, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRAD Sequencing - Oly Oyster gDNA-01 RAD Library (from 20141110)\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nNov 17, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Oly Oyster gDNA-01 RAD Library\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 10, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Prep - Oly Oyster gDNA-01 RAD\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 7, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing & Size Selection - Oly Oyster gDNA RAD P1 Adapters (from 20141105)\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 6, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLigation - Illumina P1 Adapters for Oly Oyster gDNA-01 RAD Sequencing (from 20141031)\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 5, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digest - Oly Oyster gDNA-01 for RAD Sequencing (from 20141029)\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 31, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Allocation - Oly Oyster gDNA-01 for RAD Sequencing (from 20141022)\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 29, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Oly Oyster gDNA-01 for RAD Sequencing (from 20141014)\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 22, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Olympia Oyster Populations for RAD Sequencing\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 14, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPackage - Received Package from Jerome LaPeyre from LSU\n\n\n\n\n\n\n\n2014\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 26, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas Larvae from Katie Latterhos\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 23, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas Larvae from Katie Latterhos and Emma\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 22, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Mac’s Bisulfite-Treated DNA\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 3, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Mac’s Bisulfite-Treated DNA\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 28, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA-Seq - Sea Star Data Download\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nMay 28, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Jessica’s Geoduck Larval Stages\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Mackenzie’s C.gigas EE2 Gonad Samples\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Colleen Sea Star (Pycnopodia) Coelomycete RNA for Illumina Sequencing\n\n\n\n\n\n\n\n2014\n\n\nSamples Submitted\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Colleen Sea Star (Pycnopodia) Coelomycete Sample\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Colleen Sea Star (Pycnopodia) Coelomycete Samples\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 28, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Claire’s C.gigas Female Gonad\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nApr 25, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation - Colleen’s Sea Star Coelomycete RNA from Yesterday\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 25, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Clean Up - Colleen’s Sea Star Coelomycete RNA from 20140416\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 24, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Colleens’ Sea Star Coelomycetes Samples\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 16, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Test Sample\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 16, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPhenol-Chloroform DNA Clean Up - Mac and Claire’s Samples (from 20140410)\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Sea Star Coelomocytes (from Colleen)\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Claire’s C.gigas Female Gonad and Mac’s C.gigas Gonad\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nCloned Hard Drive - Windows XP Opticon Computer (Aquacul8)\n\n\n\n\n\n\n\n2014\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nApr 4, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA gel - Claire’s C.gigas Female Gonad and Mac’s C.gigas Gonad\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nApr 4, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Sea Star Coelomocytes (provided by Colleen Burge)\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 2, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Sea Star Coelomocytes (provided by Colleen Burge)\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 1, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Mackenzie’s C.gigas Gonad Sample\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 1, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas Female Gonads (from frozen)\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nMar 28, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quality Check - Yanouk’s Oyster gDNA\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 19, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Precipitation - Geoduck DNA from 20140213\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 18, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 13, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 12, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGylcogen Assay - Emma’s C.gigas Whole Body Samples (continued from yesterday)\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 7, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGylcogen Assay - Emma’s C.gigas Whole Body Samples\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 6, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGylcogen & Carboyhydrate Assays - Emma’s C.gigas Whole Body Samples (continued from yesterday)\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 26, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGylcogen and Carboyhydrate Assays - Emma’s C.gigas Whole Body Samples\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Lake Trout C1q\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 28, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Lake Trout C1q\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 10, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Yanouk’s DNA Samples\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 4, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Lake Trout C1q\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 19, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Hexokinase Partial CDS\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Hexokinase and Partial Exon #1\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Hexokinase Promoter and CDS (repeat from 20130227)\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Hexokinase Promoter and CDS\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 27, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Herring RNA from 20091026\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 13, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Halley cDNA Check\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 28, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - FISH441 RNA\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 21, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Manila Clam Larvae cDNA (from August 2012 - Dave’s Notebook)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 8, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Manila Clam Larvae cDNA (from August 2012 - Dave’s Notebook)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 8, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Manila Clam Larvae cDNA (from August 2012 - Dave’s Notebook)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 8, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased Manila Clam Larvae RNA (from August 2012 - Dave’s Notebook)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 26, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReceived oysters from Taylor Shellfish.\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 16, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Opticon Test\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 10, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMinipreps - Emma’s Illumina Library Cloning\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nIllumina RNAseq Library Construction - 32 C.gigas Individuals\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nOligo Reconstitution - Illumina RNAseq Library Oligos and Barcodes\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nChloroform Clean Up - Lexie’s QPX RNA from 20110504\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - QPX RNA and DNA for Illumina 36bp single-end RNA/DNAseq\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nQPX Sample Pooling for Illumina Sequencing\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Detection of V.tubiashii Presence and Expression Using VtpA Primers in DNA/cDNA from yesterday\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 1, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - DNased C.gigas Larval RNA from 20120427\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\nM-MLV\n\n\nreverse transcription\n\n\nRNA\n\n\nVibrio tubiashii\n\n\n\n\n\n\n\n\n\n\n\nApr 30, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Check DNased RNA from Earlier Today for Residual gDNA\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 27, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - C.gigas Larvae RNA from yesterday\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 27, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - C.gigas Larvae from Taylor Summer 2011\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - C.gigas Larvae from Taylor Summer 2011\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Taylor Water Filter DNA Extracts from 20120322 - Sam White\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 26, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Repeat of qPCR from Earlier Today\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 23, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Taylor Water Filter DNA Extracts from Yesterday\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 23, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Extraction - Taylor Water Filter Samples from 2011\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 22, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Dave’s Manila Clam (Venerupis philippinarum) DNased RNA from 20120307 and 20120302\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 12, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Dave’s Manila Calm (Venerupis philippinarum) DNased RNA from yesterday and 20120302\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 8, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Dave’s Manila Clam (Venerupis philippinarum) Gill Samples (#25-48)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 7, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - Dave’s Manila Clam (Venerupis philippinarum) Gill RNA from Yesterday\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Dave’s Manila Clam (Venerupis philippinarum) Gill Samples (#1-24)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - cDNA from 20120208\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - cDNA from earlier today\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 8, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - C.gigas larvae DNased RNA (from 20120125)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 8, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased RNA from earlier today\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 25, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNAse - C.gigas RNA from 20120124\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 25, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - C.gigas Larvae from 20110412 & 20110705 (Continued from 20120112)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 24, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - C.gigas Larvae from 20110412 & 20110705\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - COX/PGS Clones from yesterday/today\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 14, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMini-preps - COX/PGS Cloning Colonies from today\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 13, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - COX/PGS Cloning Colony Screens from yesterday\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 13, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nCloning - Purified COX/PGS “qPCR Fragment” from 20111006\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 12, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Purified COX/PGS 1/2 DNA from earlier today\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 6, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Region Outside of COX/PGS qPCR Primers\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 6, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation - Full-length PGS1 cDNA (from 20110921)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 29, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Full-length PGS1 cDNA\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 21, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation - Purified PGS1 PCR from yesterday\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 21, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Full-length PGS1 cDNA\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 20, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Full-length PGS2 cDNA\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 14, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Full-length PGS1 & PGS2 cDNAs\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas V.vulnificus Exposure cDNA (from 20110311)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - C.gigas COX2/PGS2 Clone #4 from 20110728\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 4, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPlasmid Isolation & Sequencing - C.gigas COX2/PGS2 Clones (from yesterday)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 28, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacterial Cultures - C.gigas COX2/PGS2 Clones (from yesterday)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 27, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nColony PCRs - C.gigas COX2/PGS2 Clones (from yesterday)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 26, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nCloning - C.gigas COX2/PGS2 5’/3’ RACE Products (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\n5’/3’ RACE - C.gigas COX2/PGS2 Nested RACE PCR\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\n5’/3’ RACE - C.gigas COX2/PGS2 RACE PCR\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 22, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - PGS Hi 4 (PGS2/COX2)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 15, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPlasmid Isolation - Miniprep on PGS Hi 4 Colony from yesterday\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 12, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Colony PCR on Restreaked PGS2 Clones from 20110707\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacterial Cultures - Liquid Cultures of PGS2/COX2 Colonies from 20110707\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nClone Restreaking - PGS2 Hi/Lo Clones (from 20110421)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 7, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas GAPDH second rep on V.vulnificus exposure cDNA (from 20110311) and standard curves for COX1, COX2, GAPDH\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 3, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas actin and GAPDH on V.vulnificus exposure cDNA (from 20110311)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 2, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas 18s and EF1a on V.vulnificus exposure cDNA (from 20110311)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 2, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas COX2 on V.vulnificus exposure cDNA (from 20110311)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas COX1 on V.vulnificus exposure cDNA (from 20110311)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Hard Clam NGS Primer Checks\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Lexie’s QPX Temp & Tissue Experiment (see Lexies Notebook 4/26/2011)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 20, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Emma’s New 3KDSqPCR Primers\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 20, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Hard Clam Gill DNased RNA (from 20110509)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - Hard Clam Gill RNA (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 9, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Hard Clam Gill Tissue from Vibrio Experiment (see Dave’s Notebook 5/2/2011)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 9, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPrimer Design - Hard Clam NGS Primers\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Hard Clam Gill Tissue from Vibrio Experiment (see Dave’s Notebook 5/2/2011)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 6, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReceived - Live Hard Clams From Scott Lindell\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 28, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacterial Cultures - Colonies Selected from Yesterday’s Colony PCRs\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMini Preps - Liquid Cultures from yesterday\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacterial Cultures - Colonies Selected by Steven from Steven’s Re-Streaked Plate\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nColony PCR - Colonies from COX1 Genomic Cloning (from 20110411)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nColony PCR - 5’RACE Colony: COX2 (repeat of yesterday’s PCR)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 22, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nColony PCR - 5’ RACE Colony: COX2\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 21, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nColony PCR - 5’ RACE Colonies\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLigations - COX1/COX2 PCR Products\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\n5’/3’ RACE PCRs - Nested PCRs for COX2 Sequence\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 7, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\n5’/3’ RACE PCRs - COX2 Sequence on 5’ & 3’ RACE Libraries (from 20080619)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 6, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas BB/DH cDNA for PROPS (TIMP3(BB) primers)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMar 21, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas BB/DH cDNA for PROPS (HMGP primers)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMar 21, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas COX1/COX2 Tissue Distribution\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 15, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas BB/DH cDNA for PROPS\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - C.gigas BB/DH DNased RNA (from 20090507) for PROPS\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMar 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - C.gigas DNased RNA (from 20110131) from V.vulnificus Exposure & Tissues (from 20110111)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD Sequencing Submission\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 10, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Pooled Black Abalone Dg RNA (from Abalone Dg Exp 1)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 7, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\n3’RACE - C.gigas 3’RACE for COX2\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 4, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoDrop1000 Comparison - Roberts vs. Young Lab\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Check DNased RNA BB01 for Residual gDNA (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - C.gigas BB01 (PROPS) RNA (from 20090507)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Check DNased RNA BB01 for Residual gDNA (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - C.gigas BB01 from 20110225\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precpitation - DNased RNA BB01 (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Check DNased RNA BB01 for Residual gDNA (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - C.gigas BB01 from 20110216\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation - DNased RNA BB01 (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 16, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Check DNased RNA BB01 & 09 (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 16, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - C.gigas BB/DH (PROPS) RNA (from 20090507)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 16, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Young Lab ABI 7300 Calibration Checks\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Test Young Lab qPCR Calibration\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoDrop1000 Comparison - Roberts vs. Young Lab\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Test Young Lab qPCR Calibration (Repeat)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 8, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - New C. gigas COX Primers for Sequencing of Isoforms\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 8, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Test Young Lab qPCR Calibration (Repeat)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 8, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Test Young Lab qPCR Calibration (Repeat)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 4, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Test Young Lab qPCR Calibration\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 4, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic PCR - Repeat of C.gigas COX genomic PCR from 20110118\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 31, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - DNase C.gigas RNA from 20110120, 20110121 and 20110124\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 28, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Various C.gigas Tissue from 20110111\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 24, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Various C.gigas Tissue from 20110111\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 21, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Various C.gigas Tissue from 20110111\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 20, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic PCR - C.gigas cyclooxygenase (COX) genomic sequence\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 18, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacterial Dilutions - Determination of Colony Forming Units from Gigas Bacterial challenge (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGigas Bacterial Challenge - 1hr & 3hr Challenges with Vibrio vulnificus\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD - Retrieved SOLiD Library Samples from CEG from 20101213\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 23, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - COX qPCR Vibrio Exposure Response Check\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 13, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - COX qPCR Primer Test and Tissue Distribution\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 10, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digestions/Ligations - MS-AFLP\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digestions - HpaII and MspI on Mac’s C.gigas Samples: Round 1\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 29, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEtOH Precipitations - HpaII and MspI 2nd Round Digests from 20101124\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 29, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digestions - HpaII and MspI on Mac’s C.gigas Samples: Round 2\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPhenol:Chloroform Extractions and EtOH Precipitations - HapII and MspI digests from yesterday\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 23, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digestions - HpaII and MspI on Mac’s C.gigas gDNA Samples: Round 1\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 22, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nQPX Washes\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 26, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 26, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEtOH Precipitation - Whale gDNA from 20101022\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 25, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 22, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReceived Hard Clam Samples and Live Clams from MBL\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 19, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReceived Hard Clam Samples from Rutgers\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 15, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Test Plate for Opticon 2\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 12, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nOpticon Calibration\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Hard Clam Primers on cDNA from yesterday\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\nBigDef\n\n\nC1qTNG\n\n\nCA\n\n\nCFX96\n\n\nCytP450-like\n\n\nferritnin\n\n\ngraphs\n\n\nGST\n\n\nHard clam\n\n\nHemoDef\n\n\nHSP 70\n\n\nImmomix\n\n\nlysozyme\n\n\nMA\n\n\nMAX\n\n\nMercenaria mercenaria\n\n\nMercenaria_Rel\n\n\nmetallothionein\n\n\nMm_TRAF6\n\n\nNADHIV\n\n\nqPCR\n\n\nRACK1\n\n\nSenescentProt\n\n\nSTI\n\n\nSYTO 13\n\n\nTfAP1\n\n\nTLR\n\n\n\n\n\n\n\n\n\n\n\nSep 9, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - DNased Hard Clam RNA from earlier today\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - DNasing Hard Clam RNA from yesterday\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEtOH Precipitation - Hard Clam RNA from earlier today\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 7, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Hard Clam Tissues Rec’d from Rutgers on 20100820\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 7, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPackage - Hard Clam Samples from MBL\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 2, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPackage - Hard Clam Samples from Rutgers\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 20, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - HpaII/MspI Digests from earlier today\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digests - Various gigas gDNA from earlier today\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Various gigas samples (continued from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Various gigas samples\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMeDIP - SB/WB Fragmented gDNA EtOH precipitation (continued from 20100702)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMeDIP - SB/WB Fragmented gDNA (continued from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digests - Various gigas gDNAs of Mac’s\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMeDIP - SB/WB Fragmented gDNA (continued from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 1, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMeDIP - SB/WB Fragmented gDNA (from 20100625)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - Fragmented SB/WB gDNA (from 20100625)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Sonication - SB/WB gDNA pools (prep for MeDIP) from 20100618\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Hard Clam samples from Rutgers and MBL\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 24, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Sonication - SB/WB gDNA pools (prep for MeDIP) from earlier today\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 18, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Precipitation - SB/WB gDNA pools (prep for MeDIP)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 18, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Mac gigas larvae samples: control larvae 6.7.10 and 5-aza tr larvae 6.7.10\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Mac gigas gill samples (continued from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Mac gigas gill samples\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\nCrassostrea gigas\n\n\nDNA Isolation\n\n\nDNazol\n\n\ngill\n\n\nPacific oyster\n\n\nR37\n\n\nR51\n\n\nRNAlater\n\n\n\n\n\n\n\n\n\n\n\nJun 4, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Mac gigas gill samples (continued from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 28, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPackage Rec’d - From NOAA in Connecticut\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\nalgae\n\n\nIsochrysis sp. T-150\n\n\nNOAA\n\n\npackage\n\n\nTetraselmis cheri Ply429\n\n\nThalassiaosira weissflugii TW\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Mac gigas gill samples\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\nCrassostrea gigas\n\n\nDNazol\n\n\nPacific oyster\n\n\nR51\n\n\nRNAlater\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD ePCR/Templated Bead Prep - Lake Trout Lean library\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 26, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD Templated Bead Prep - Yellow perch CT, WB and lake trout Lean libraries (continued from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 13, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD ePCRs - Yellow perch CT, WB and lake trout Lean libraries\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 12, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPackage - Hard Clam gill tissue/hemolymph in RNA later\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 5, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Qiagen Kit Comparison\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\nDNeasy\n\n\ngDNA\n\n\ngel\n\n\nHyperladder I\n\n\ntroubleshooting\n\n\n\n\n\n\n\n\n\n\n\nMay 5, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - V.tubiashii primers test (Vpt A and Vt IGS)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 3, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTemplated Bead Prep SOLiD Libraries - Yellow perch WB, lake trout Lean and Sisco, and herring G/O HWS09 libraries\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 16, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nePCR SOLiD Libraries - Lake Trout Sisco and Herring G/O HPWS09 libraries (from 20100408)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTemplated Bead Prep SOLiD Libraries - Abalone CC, CE pools and yellow perch CT, PQ libraries\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nePCR SOLiD Libraries - Yellow perch PQ, WB and Lake Trout Lean libraries (from 20100408)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nePCR SOLiD Libraries - Abalone CC, CE pools and yellow perch CT SOLiD libraries (from 20100408)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 12, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ncDNA clean up & Bioanalyzer for SOLiD Libraries - Abalone, Yellow Perch, Lake Trout, Herring\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGel Purification & PCR cDNA SOLiD Libraries - Abalone, Yellow Perch, Lake Trout, Herring\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 7, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription SOLiD Libraries - Abalone, Yellow Perch, Lake Trout, Herring\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 6, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHibridizaton/Ligation SOLiD Libraries - Abalone, Yellow Perch, Lake Trout, Herring\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 5, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer Total, mRNA and post-fragmentation SOLiD Libraries - Abalone pools\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 2, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation and Fragmentation for SOLiD Libraries - Pooled abalone mRNA (from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 1, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation & mRNA Isolation for SOLiD Libraries - Pooled abalone total RNA: Carmel control, Carmel exposed\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 31, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer for SOLiD Libraries - Fragmented mRNA from Perch, Lake Trout & Herring RNA samples\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 29, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD Library Prep - mRNA (perch, lake trout, herring from 20100318) Fragmentation\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 25, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer for SOLiD libraries - Total and mRNA from Perch, Lake Trout & Herring RNA samples (CONTINUED from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Precipitation for SOLiD - Perch, Lake Trout, & Herring mRNA (CONTINUED from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 17, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation for SOLiD - Perch, Lake Trout, and Herring total RNA\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 16, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Test Lexie’s Mercenaria 18s contamination issue\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 10, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Mac’s BB/DH cDNA from 20091223\n\n\n\n\n\n\n\n2010\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nJan 15, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD Bead Titration - Herring fragmented cDNA libraries: 2LHKOD09, 4LHTOG09, 6LHPWS09\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD ePCRs - Herring cDNA libraries\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 13, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD ePCR - Herring fragmented cDNA library: 2LHKOD09\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - Mac’s BB/DH cDNA from 20091223\n\n\n\n\n\n\n\n2010\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD Bead Titration - Herring fragmented cDNA library 3LHSITK09 (CONTINUED from ePCR yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD ePCR - Herring fragmented cDNA library: 3LHSITK09\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 7, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD Bead Titration - Herring fragmented cDNA library 3LHSITK09(CONTINUED from ePCR yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 7, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - Tim’s Adult Gigas gill cDNA (from 20091009)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 6, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD ePCR - Herring fragmented cDNA library: 3LHSITK09 (from 20091209)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 6, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - Tim’s Adult Gigas gill cDNA (from 20091009)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 5, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - BB & DH cDNA (from 20091223)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 31, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - BB & DH cDNA (from 20091223)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 30, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - BB & DH cDNA (from 20091223)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAlaska sockeye salmon sampling (with Seebs): Family #13\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - BB & DH cDNA (from 20091223) and Emma primer sets for testing\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - BB & DH cDNA (from 20091223)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 28, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - BB & DH cDNA (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - Mac methylation samples, Sam rhodopsin samples, Lisa samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - BB & DH cDNA (from earlier today)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - BB & DH DNased RNA (from 20090514)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Sepia cDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Sepia cDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Sepia cDNA and DNased RNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Sepia cDNA (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Abalone 07:12 DNased RNA (from 20090623)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Sepia DNased RNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - Herring Liver cDNA for SOLiD Libraries\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEmulsion PCR - Herring Liver cDNA for SOLiD Libraries\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Herring Liver mRNA for SOLiD Libraries\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Adapter Hybridization and Ligation - Herring Liver mRNA for SOLiD Libraries\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Fragmentation - Herring Liver mRNA for SOLiD Libraries\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Sepia samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 4, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - Dungan Isolates, Lake Trout HRM and Emma DD cloning\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 3, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Precipitation - Herring Liver mRNA for SOLiD Libraries (continued from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 3, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHard Clam Challenge - QPX Strain S-1 (continued from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation - Herring Liver RNA for SOLiD Libraries (continued from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation - Herring Liver RNA for SOLiD Libraries\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHard Clam Challenge - QPX Strain S-1\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBL Shipment - Hard Clam gill tissue in RNA Later\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBL Shipment - Sepia tissue samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBL Shipment - Hard Clams\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHerring 454 Data\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 19, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBL Shipment - MV oysters/cod\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 19, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - “Unknown” Dungans/Lyons\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 18, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - “Unkown” Dungans/Lyons\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 17, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHard Clams - Shipment from Rutgers\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - Lake Trout HRM\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nOyster CO2/Mechanical Stress - Water quality\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submissions to MoGene for 454 Analysis - Herring Liver and Testes mRNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nOct 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Herring gonad/ovary RNA (from 20091023)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation - Herring gonad/ovary RNA (from 20091023)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 26, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Herring Liver RNA (from 20091021)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 26, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation - Herring Liver Samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Herring Gonad/Ovary Samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Herring Gonad/Ovary Samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Herring Liver Samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 21, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Herring Liver Samples (LHPWS09 1-6)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 20, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - Tim’s adult gigas challenge cDNA (from 20091009)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Tim’s adult gigas challenge cDNA (from 20091009)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Tim’s adult gigas challenge cDNA (from today)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Tim’s adult gigas challenge DNased RNA (from 20091008)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Tim’s adults gigas challenge re-DNased RNA (from today)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Re-DNase of Tim’s adult gigas challenge RNA (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Tim’s adults gigas challenge DNased RNA (from today)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Tim’s adults gigas challenge DNased RNA (from today)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Tim’s adult gigas challenge RNA (from 20090930)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer Submission - Trout RBC, Colleen’s gigas GE sample, Mac’s DH/BB PCR for SOLiD WTK\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription/cDNA purification/Emulsion PCR - Ligation rxns of trout fragmented RNA for SOLiD WTK (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAdapter Ligation - Rick’s trout fragmented control/poly I:C samples for SOLiD WTK\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Tim’s adults gigas challenge DNased RNA (from 20091002)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Tim’s adult gigas challenge RNA (from 20090930)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - Check gDNA contamination with EF1 & 18s primers in gigas gill RNA (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Tim’s adult gigas challenge samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 30, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer Submission - Rick’s trout RBC samples (various dates)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Fragmentation - Rick’s trout RBC samples prepped earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEtOH Precipitation - Rick’s trout Ribosomoal-depleted RNA for SOLiD WTK (from today)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRibosomal-depleted RNA - Rick’s trout RBC samples for the SOLiD WTK\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Fragmentation - Rick’s trout RBC samples prepped earlier today (see below)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Rick’s trout RBC samples previously treated with Ribominus Kit (by Mac)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Gigas BB and DH samples previously treated with Ribominus Kit (by Mac)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Gigas BB and DH samples previously treated with Ribominus Kit (by Mac)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHRMs - Lake Trout SNPs (HRM_white-05 & HRM_white_06)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHRMs - Lake Trout SNPs (HRM_white-03 & HRM_white-04)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHRM - Lake Trout SNPs (HRM_white-02)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHRM - Lake Trout SNPs (HRM-white-01)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 3, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - HRM Lake Trout SNP primer test\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPrimers - Lake Trout Primers for HRM\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Gigas gDNA test of recalibrated Opticon 2\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Recalibration of Opticon 2\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 26, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Precipitation - C.pugetti DNA for JGI submission (continued from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 26, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Precipitation - C.pugetti DNA for JGI submission\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 25, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Additional Calibration test of Opticon 2\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Carita Primer Test for High Resolution Melt (HRM) Curve Analysis\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 17, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Calibration test of Opticon 2\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Calibration test of Opticon 2\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C.pugetti culture (from 20090713)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Gigas DNA for Opticon testing\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRT Rxns - H.crach DNased RNA (from 20090623)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoDrop - H.crach DNased RNA (from 20090623)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 21, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone cDNA (07:12 set from 3/3/2009 by Lisa) and DNased RNA (from 20090623)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 20, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone gDNA (H.crach 06:7-1)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 17, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone cDNA (07:12 set from 3/3/2009 by Lisa) and DNased RNA (from 20090623)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone cDNA (07:12 set from 3/3/2009 by Lisa) and DNased RNA (from 20090623)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone gDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone RNA/DNased RNA & “dirty” and “clean” cDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Dungan isolate (MIE-14v) gDNA from 20090708\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C.pugetti large culture\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone gDNA/cDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased Abalone Dg RNA from 20090625\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Precipitation CONTINUED - Dungan MIE-14v gDNA from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Bay/Sea Scallop DNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased Abalone Dg RNA from 20090625\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Precipitation - Dungan MIE-14v gDNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Dungan isolate MIE-14v\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSpec Reading - C.pugetti gDNA from 20090526\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti liquid cultures\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 26, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone Dg DNased RNA from yesterday and earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg RNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg RNA (07:12 set)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEtOH Precipitation - DNased Abalone Dg RNA from yesterday AND the 07:12 set (DNased by Lisa 20090306)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased Abalone Dg RNA from earlier today AND the 07:12 set (DNased by Lisa 20090306)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg DNased RNA 20090610\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 18, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 17, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA: Test Immomix (SYTO13) vs. Strategene SYBR\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - MV hemocyte DNased RNA from 20090612\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased MV hemocyte RNA from earlier today AND Turbo kit test\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - MV hemocyte RNA from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - MV hemocyte RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 11, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Martha’s Vineyard (MV) hemocytes\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 11, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Re-DNased abalone Dg RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Re-DNased abalone Dg RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg DNased RNA from 20090605\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - C.pugetti gDNA from 20090526\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - C.pugetti gDNA from 20090513 & 20090526\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Re-DNased abalone Dg RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg DNased RNA from 20090605\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C.pugetti culture (1x 1L)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased abalone Dg RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg RNA isolated yesterday and from 20090518\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Abalone Dg Project Samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 4, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg RNA isolated yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 3, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - C.pugetti DNA from 20090513 & 20090526\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Abalone Dg Project\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - C.pugetti DNA from 20090513 & 20090526\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Gel - JGI QC check of C. pugetti DNA from 20090526\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - C. pugetti (from 20090518)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 26, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Test - Gigas site gDNA (BB & DH) from 20090515\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Abalone Dg Project samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 18, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nC.pugetti - Liquid Cultures\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 18, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Gigas Dermo Samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Mac’s BB and DH site samples\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Mac’s gigas DNased RNA from 20090512\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - C.pugetti\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Mac’s gigas DNased RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment (Rigorous!) - Mac’s gigas RNA/Re-DNased RNA from 20090507 & 20090508, respectively\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Mac’s gigas samples from 20090505 & 20090506\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 11, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Re-DNased oyster RNA from today\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Oyster RNA from yesterday\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased oyster RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Oyster RNA from today\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Mac’s oyster tissues (BB and DH) (CONTINUED from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Mac’s oyster tissues (BB and DH) (CONTINUED from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti liquid cultures\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Mac’s oyster tissues (BB and DH)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti liquid cultures\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti plate (from 20090424)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 30, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti plate (from 20090424)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti liquid culture\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti plate\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - cDNA from DNased Abalone RNA from 20090420\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone DNased RNA from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 21, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugettii culture CONTINUED (from 20090419)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 21, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Removal - Abalone RNA from 20090402 and 20090331\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 20, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugettii culture\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 19, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - Dungan isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 17, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Two new Dungan isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Rab7_SYBR primers on abalone RNA and DNased RNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Two new Dungan isolates from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Two new Dungan isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Test QT Kit with No RT Abalone rxns from 20090408\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Bay/Sea scallop gDNAs\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Bay/Sea scallop gDNA isolated earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Bay/Sea Scallop and hybrid samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Abalone gDNA/RNA/cDNA w/new TOLLIP primer\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Old Dungan isolates #1-35 w/EukA/B primers\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Bay/Sea scallop hybrids\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Dungan isolates from 20090402 with Euk primers\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Check DNased abalone RNA (by Lisa) for gDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Repeat (modified) of yesterday’s abalone cDNA check\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone cDNA (QT) from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ncDNA - Abalone RNA from 20090331 & 20090402\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - New Dungan isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - New Dungan isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 3, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone RNA, check for gDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 3, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - New Dungan isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Abalone digestive gland samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Abalone digestive gland samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 31, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Abalone digestive gland samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Virginica cDNA (see workup sheet below for more info regarding cDNA)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Sample Submission Hard clam gill #1 mRNA from 20090313\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - hard clam gill #1 continued from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - hard clam gill #1 DNased RNA from today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation - Hard clam gill #1 RNA from 20080819\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Repeat of qPCR from earlier today with fresh primer working stocks\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Repeat of 20090227 qPCR with clean water\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - New 16s primers for V.tubiashii Control vs. Autoclaved gigas samples (see 20090224)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Replicate of V.tubiashii Control vs. Autoclaved gigas samples (see yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 25, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - V.tubiashii Control vs. Autoclaved gigas samples (see below)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - V.tubiashii DNAsed RNA (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - V.tubiashii samples from autoclaved gigas exposure (from 20081218)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEpigenetics Experiment - Gigas treatment\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 20, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - V.tubiashii Mass Spec\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 18, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrypsin digestion - Vibrio 2D spots CONTINUED (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrypsin digestion - Vibrio 2D spots from 20081217\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 11, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA - Submission for Agilent Bioanalyzer\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA - Precipitation continued from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Hard Clam gill and hemo RNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA - Hard clam hemo RNA (from 20090121)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Hard clam hemo (from 20090121)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 21, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBleeding/Tissue Collection - Hard Clams\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 20, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA - Precipitation of Hard Clam Hemo RNA from 20090116\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 20, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Hard clam gill, hemos\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBleeding - Hard Clams\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA - Precipitation continued from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA - Reprecipitation of hard clam RNA from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBleeding - Hard Clams\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Hard Clam hemolymph from 20090108, 20090109\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBleeding - Hard Clams\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBleeding - Hard Clams\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Dungan Isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Dungan Isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE, Western Blot - Test of HSP70 Ab on heat stressed shellfish for FISH441\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE, Western Blot - Test of new Western Breeze Kit & HSP70 Ab for FISH441\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\nanti-HSP70\n\n\nanti-myc\n\n\nantibody\n\n\nCoomassie\n\n\nCrassostrea gigas\n\n\nMSTN\n\n\nMSTN1b\n\n\nmyostatin\n\n\nPacific oyster\n\n\nprotein\n\n\nSDS-PAGE\n\n\nSeeBlue Plus\n\n\nWestern blot\n\n\nWestern Breeze Chromogenic (anti-mouse) Kit\n\n\n\n\n\n\n\n\n\n\n\nDec 31, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Gel - V. tubiashii mRNA samples (from 20081224)\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 29, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nrRNA Removal - V. tubiashii total RNA from yesterday\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 24, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - V. tubiashii from challenge (see 20081216)\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\nMICROBExpress Kit\n\n\nNanoDrop1000\n\n\nRNA isolation\n\n\nRNA quantification\n\n\nTriReagent\n\n\nVibrio tubiashii\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nVibrio challenge CONTINUED (from yesterday)\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 19, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nVibrio challenge CONTINUED (from yesterday)\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nVibrio challenge CONTINUED (from yesterday)\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nVibrio challenge\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\nbacterial challenge\n\n\nbacterial culture\n\n\nVibrio exposure\n\n\nVibrio tubiashii\n\n\n\n\n\n\n\n\n\n\n\nDec 16, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE/Western - anti-HSP70 Ab Re-test\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\nanti-HSP70\n\n\nantibody\n\n\nchemiluminescent\n\n\nCoomassie\n\n\nCrassostrea gigas\n\n\nhemocyte\n\n\nhemolymph\n\n\nPacific oyster\n\n\nprotein\n\n\nSDS-PAGE\n\n\nSeeBlue Plus\n\n\n\n\n\n\n\n\n\n\n\nDec 16, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE/Western - Attempt to fix/identify problem(s) with Westerns\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 12, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE/Western - anti-HSP70 Ab test CONTINUED (from yesterday)\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 10, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE/Western - anti-HSP70 Ab test\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\nanti-HSP70\n\n\nantibody\n\n\nCoomassie\n\n\nCrassostrea gigas\n\n\ngill\n\n\nmucus\n\n\nOctopus rubescans\n\n\nPacific oyster\n\n\nprotein\n\n\nred octopus\n\n\nSDS-PAGE\n\n\nskin\n\n\n\n\n\n\n\n\n\n\n\nDec 9, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE/Western - Purified (His column) FST samples from 20081112\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 17, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - Y2H colony PCRs from 20081112\n\n\n\n\n\n\n\n2008\n\n\nMyostatin Interacting Proteins\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMass Spec - Band #1 from 20081106\n\n\n\n\n\n\n\n2008\n\n\nMyostatin Interacting Proteins\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nWestern Blot - Purified (His column) decorin, FST, LAP & telethonin\n\n\n\n\n\n\n\n2008\n\n\nMyostatin Interacting Proteins\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2008\n\n\nSam White\n\n\n\n\n\n\nNo matching items"
+ "text": "Data Exploration - CEABIGR Spurious Transcription Calculations and Plotting\n\n\n\n\n\n\n\n2024\n\n\nCEABIGR\n\n\nplot\n\n\nEastern oyster\n\n\nCrassostrea virginica\n\n\n\n\n\n\n\n\n\n\n\nJan 3, 2024\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - January 2024\n\n\n\n\n\n\n\n2024\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nJan 2, 2024\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputer Management - Returned Failing HDDs from Gannet Expansion Unit\n\n\n\n\n\n\n\ncomputer management\n\n\n2023\n\n\nHDDs\n\n\ngannet\n\n\n\n\n\n\n\n\n\n\n\nDec 21, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Gadus macrocephalus liver tissues to Azenta for RNA-seq\n\n\n\n\n\n\n\n2023\n\n\nGadus macrocephalus\n\n\nPacific cod\n\n\nAzenta\n\n\nRNA-seq\n\n\nSamples Submitted\n\n\nliver\n\n\nspleen\n\n\ngill\n\n\nblood\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputer Maintenance - Replace Failing HDDs in Synology RX1217 Expansion Unit on Gannet\n\n\n\n\n\n\n\ngannet\n\n\nsynology\n\n\nRX1217\n\n\nHDDs\n\n\n2023\n\n\ncomputer maintenance\n\n\n\n\n\n\n\n\n\n\n\nDec 7, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Investigation - Mysterious Case of Excessive Exons in CEABIGR\n\n\n\n\n\n\n\nCEABIGR\n\n\nCrassostrea viriginica\n\n\nEastern oyster\n\n\n2023\n\n\n\n\n\n\n\n\n\n\n\nDec 6, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - December 2023\n\n\n\n\n\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPlotting Fun - Lab Meeting Task\n\n\n\n\n\n\n\nplotting\n\n\nR\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - M.magister De Novo Transcriptome Assembly Using Trinotate on Mox\n\n\n\n\n\n\n\nmox\n\n\n2023\n\n\nMetacarcinus magister\n\n\nDungenss crab\n\n\nTrinotate\n\n\ntranscriptome\n\n\nannotation\n\n\n\n\n\n\n\n\n\n\n\nNov 9, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - M.magister De Novo Transcriptome Assembly for DuMOAR Project Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\nmox\n\n\nDIAMOND\n\n\nBLASTx\n\n\ntranscriptome\n\n\n2023\n\n\nDuMOAR\n\n\nDungeness crab\n\n\nMetacarcinus magister\n\n\n\n\n\n\n\n\n\n\n\nNov 9, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - November 2023\n\n\n\n\n\n\n\nDaily Bits\n\n\n2023\n\n\n\n\n\n\n\n\n\n\n\nNov 5, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR Analysis - C.gigas Matt George Poly:IC Diploids\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 22, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Identification and Alignments - C.virginica RNAseq with NCBI Genome GCF_002022765.2 Using Hisat2 and Stringtie on Mox Again\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 21, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas PolyIC Diploid MgCl2\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 17, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - C.gigas PolyIC Diploid MgCl2\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Quantification - C.gigas PolyIC Diploid MgCl2\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - August 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nAug 1, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nsRNA-seq Alignments - E5 Coral P.evermanni Using ShortStack on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Extractions - M.magister MEGAN FastA to FastQ Arthropoda and Unassigned Reads\n\n\n\n\n\n\n\n2023\n\n\nDuMOAR\n\n\n\n\n\n\n\n\n\n\n\nJul 30, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDIAMOND BLASTx - C.virginica Genes on Mox\n\n\n\n\n\n\n\n2023\n\n\nCEABIGR\n\n\n\n\n\n\n\n\n\n\n\nJul 27, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas polyIC\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.virginica Genes Only FastA from Genes BED File Using gffread on Raven\n\n\n\n\n\n\n\n2023\n\n\nCEABIGR\n\n\n\n\n\n\n\n\n\n\n\nJul 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Extractions - M.magister MEGAN Arthropoda and Unassigned Reads to FastA\n\n\n\n\n\n\n\n2023\n\n\nDuMOAR\n\n\n\n\n\n\n\n\n\n\n\nJul 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Quantification - C.gigas PolyIC\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 21, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - C.gigas PolyIC RNA\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 21, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas Ctenidia cDNA for Noah’s Heat-Mechanical Stress Project\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 21, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas Ctenidia cDNA for Noah’s Heat-Mechanical Stress Project\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 19, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas cDNA Primer Tests for Noah’s Heat-Mechanical Stress Project\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 13, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - C.gigas RNA from Noah’s Heat-Mechanical Stress Project\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 13, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Quantification - C.gigas RNA from Noah’s Heat-Mechanical Stress Project\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 13, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - C.gigas Ctenidia from Noah’s Heat-Mechanical Stress Project\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 12, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - July 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nJul 5, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFile Conversion - M.magister MEGANized DAA to RMA6\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Pacific Cod (G.macrocephalus) Sequencing Data from Novogene\n\n\n\n\n\n\n\n2023\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nsRNA-seq Alignments - E5 Coral A.pulchra P.meandrina Using ShortStack on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nJun 28, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming and QC - E5 Coral sRNA-seq Data fro A.pulchra P.evermanni and P.meandrina Using FastQC flexbar and MultiQC on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nJun 20, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nORF Identification - L.staminea De Novo Transcriptome Assembly v1.0 Using Transdecoder on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 17, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - De Novo L.staminea Trimmed RNAseq Using Trinity on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - L.staminea RNA-seq Using FastQC fastp and MultiQC on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - June 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nJun 2, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeats Identification - P.meandrina Using RepeatMasker on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nMay 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming and QC - E5 Coral sRNA-seq Trimming Parameter Tests and Comparisons\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nMay 24, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - P.meandrina Genome GFF to GTF Using gffread\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQ QC and Trimming - E5 Coral RNA-seq Data for A.pulchra P.evermanni and P.meandrina Using FastQC fastp and MultiQC on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - E5 Coral RNA-seq and sRNA-seq Reorganizing and Renaming\n\n\n\n\n\n\n\n2023\n\n\nData Management\n\n\n\n\n\n\n\n\n\n\n\nMay 17, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Coral RNA-seq Data from Azenta Project 30-789513166\n\n\n\n\n\n\n\n2023\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Coral sRNA-seq Data from Azenta Project 30-852430235\n\n\n\n\n\n\n\n2023\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nMay 15, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nlncRNA Expression - P.generosa lncRNA Expression Using StringTie\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 4, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - May 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nMay 2, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nlncRNA Identification - P.generosa lncRNAs using CPC2 and bedtools\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 2, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nContainers - Apptainer Explorations\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 28, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Alignments - P.generosa RNA-seq Alignments for lncRNA Identification Using Hisat2 StingTie and gffcompare on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQ Trimming and QC - P.generosa RNA-seq Data from 20220323 on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - CEABIGR C.virginica Exon Expression Table\n\n\n\n\n\n\n\n2023\n\n\nCEABIGR\n\n\n\n\n\n\n\n\n\n\n\n\nApr 5, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - April 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nApr 3, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Append Gene Ontology Aspect to P.generosa Primary Annotation File\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 29, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing Read Taxonomic Classification - M.magister RNA-seq Using DIAMOND BLASTx and MEGAN6 daa-meganizer on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - March 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nMar 3, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Trimmed M.magister RNA-seq from NOAA\n\n\n\n\n\n\n\n2023\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing Read Taxonomic Classification - P.verrucosa E5 RNA-seq Using DIAMOND BLASTx and MEGAN daa-meganizer on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nFeb 21, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.goreaui Genome GFF to GTF Using gffread\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nFeb 17, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Identification and Alignments - P.verrucosa RNA-seq with Pver_genome_assembly_v1.0 Using HiSat2 and Stringtie on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nFeb 16, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQ Trimming and QC - P.verrucosa RNA-seq Data from Danielle Becker in Hollie Putnam Lab Using fastp FastQC and MultiQC on Mox\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nFeb 15, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Create P.verrucosa GCA_014529365.1 Karyotype File\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\n\n\n\n\n\n\n\n\n\nFeb 15, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - P.verrucosa RNA-seq and WGBS Full Data from Danielle Becker\n\n\n\n\n\n\n\n2023\n\n\nE5\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nFeb 10, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - February 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nFeb 1, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Indexing - P.verrucosa v1.0 Assembly with HiSat2 on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 31, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Indexing - M.capitata HIv3 Assembly with HiSat2 on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 31, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Indexing - P.acuta HIv2 Assembly with HiSat2 on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 31, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - P.verrucosa Genome GFF to GTF Using gffread\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 27, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - M.capitata Genome GFF to GTF Using gffread\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 27, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - P.acuta Genome GFF to GTF Conversion Using gffread\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 26, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Indexing - P.verrucosa NCBI GCA_014529365.1 with HiSat2 on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 25, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Indexing - M.capitata NCBI GCA_006542545.1 with HiSat2 on Mox\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 25, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Data - Coral SRA BioProject PRJNA744403 Download and QC\n\n\n\n\n\n\n\n2023\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 13, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMonthly Goals - January 2023\n\n\n\n\n\n\n\n2023\n\n\nMonthly Goals\n\n\n\n\n\n\n\n\n\n\n\nJan 5, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - January 2023\n\n\n\n\n\n\n\n2023\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nJan 3, 2023\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBS-Seq Analysis - Nextflow EpiDiverse SNP Pipeline for Haws Hawaii C.gigas BAMs from Yaamini Base Config\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 15, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBS-Seq Analysis - Nextflow EpiDiverse SNP Pipeline for Haws Hawaii C.gigas BAMs from Yaamini\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 14, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - December 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - November 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nNov 1, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - October 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nOct 1, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.virginica NCBI GCF_002022765.2 GFF to Gene and Pseudogene Combined BED File\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 26, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBSseq SNP Analysis - Nextflow EpiDiverse SNP Pipeline for C.virginica CEABIGR BSseq data\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 21, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Identify C.virginica Genes with Different Predominant Isoforms for CEABIGR\n\n\n\n\n\n\n\n2022\n\n\nCEABIGR\n\n\n\n\n\n\n\n\n\n\n\nSep 20, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Alignments - P.generosa Alignments and Alternative Transcript Identification Using Hisat2 and StringTie on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 14, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQ Trimming - Geoduck RNAseq Data Using fastp on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 8, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - September 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nSep 2, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQ Trimming and QC - C.virginica Larval BS-seq Data from Lotterhos Lab and Part of CEABIGR Project Using fastp on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 29, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Convert S.namaycush NCBI GFF to genes-only BED file for Use in Ballgown Analysis\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 18, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSplice Site Identification - S.namaycush Liver Parasitized and Non-Parasitized SRA RNAseq Using Hisat2-Stingtie with Genome GCF_016432855.1\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 10, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Repeat of Mussel Gill Heat Stress cDNA with Ferritin Primers\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 4, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - August 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nAug 3, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Dorothys Mussel cDNA from 20220726\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 27, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - Dorothy Mussel Gill Samples\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 26, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBS-seq and SNP Analysis - Nextflow EpiDiverse Pipelines Trials and Tribulations\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 25, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Data - S.namaycush SRA BioProject PRJNA674328 Download and QC\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - July 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nJul 5, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDaily Bits - June 2022\n\n\n\n\n\n\n\n2022\n\n\nDaily Bits\n\n\n\n\n\n\n\n\n\n\n\nJun 22, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Ostrea lurida MBD BS-seq from 20160203\n\n\n\n\n\n\n\n2022\n\n\nSRA Submission\n\n\n\n\n\n\n\n\n\n\n\nJun 15, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nProject Summary - C.virginica CEABiGR - Female vs. Male Gonad Exposed to OA\n\n\n\n\n\n\n\n2022\n\n\nProject Summary\n\n\n\n\n\n\n\n\n\n\n\nApr 25, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Create Primary P.generosa Genome Annotation File\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 19, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - O.nerka Berdahl Brain Tissues\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 11, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nServer Maintenance - Fix Server Certificate Authentication Issues\n\n\n\n\n\n\n\n2022\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nApr 2, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNextflow - Trials and Tribulations of Installing and Using NF-Core RNAseq\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 25, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - P.generosa Genomic Feature FastA Creation\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 24, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDifferential Gene Expression - P.generosa DGE Between Tissues Using Nextlow NF-Core RNAseq Pipeline on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 23, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - C.virginica BSseq Unmapped Reads Using MEGAN6\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 9, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - C.virginica RNAseq Zymo ZR4059 Analyzed by ZymoResearch\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTaxonomic Assignment - C.virginica BSseq Unmapped Reads Using DIAMOND BLASTx and MEGAN6 on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - P.generosa Genome GFF Conversion to GTF Using gffread\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Identification and Alignments - C.virginica RNAseq with NCBI Genome GCF_002022765.2 Using Hisat2 and Stringtie on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 25, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - Additional 20bp from C.virginica Gonad RNAseq with fastp on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 24, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.virginica lncRNA Extractions from NCBI GCF_002022765.2 Using GffRead\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 17, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Genome-guided C.virginica Adult Gonad OA RNAseq Using Trinity on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Alignment - C.virginica Adult OA Gonad Data to GCF_002022765.2 Genome Using HISAT2 on Mox\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 31, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.virginica Gonad RNAseq Transcript Counts Per Gene Per Sample Using Ballgown\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 27, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.virginica OA Larvae DNA Methylation FastQs from Lotterhos Lab\n\n\n\n\n\n\n\n2022\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nJan 19, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nProject Summary - Matt George PSMFC Mytilus Byssus Project\n\n\n\n\n\n\n\n2022\n\n\nProject Summary\n\n\n\n\n\n\n\n\n\n\n\nJan 13, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - O.nerka Berdahl Brain Tissues\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - M.trossulus Gill\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - M.trossulus Gill and Phenol Gland\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - M.trossulus Gill and Phenol Gland\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 10, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - M.trossulus Gill and Phenol Gland\n\n\n\n\n\n\n\n2022\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2022\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - M.trossulus Phenol Gland and Gill\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 23, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - M.trossulus Foot and Phenol Gland\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nProject Summary - O.nerka Berdahl Samples\n\n\n\n\n\n\n\n2021\n\n\nProject Summary\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - O.nerka Berdahl Brain Tissues\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - O.nerka Berdahl Tissues\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 20, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.virginica NCBI GCF_002022765.2 GFF to Gene BED File\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 9, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDifferential Transcript Expression - C.virginica Gonad RNAseq Using Ballgown\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 21, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSNP Characterization - C.bairdi v3.1 Transcriptome Assembly and Day2 DEG Pooled Samples RNAseq Data\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSNP Identification - C.bairdi Day 2 DEG Pooled Samples Using bcftools on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nSep 9, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Alignments - C.bairdi Day 2 Infected-Uninfected Temperature Increase-Decrease RNAseq to cbai_transcriptome_v3.1.fasta with Hisat2 on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nSep 8, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Indexing - C.bairdi Transcriptome cbai_transcriptome_v3.1.fasta with Hisat2 on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nSep 8, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputer Servicing - APC SUA2200RM2U UPS Battery Replacement\n\n\n\n\n\n\n\n2021\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nAug 24, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputer Management - Disable Sleep and Hibernation on Raven\n\n\n\n\n\n\n\n2021\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nAug 24, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nUninterruptible Power Supply - Battery Replacement for APS BR1000G\n\n\n\n\n\n\n\n2021\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nAug 11, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Identification and Quantification - C.virginia RNAseq With NCBI Genome GCF_002022765.2 Using StringTie on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 26, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotations - Splice Site and Exon Extractions for C.virginica GCF_002022765.2 Genome Using Hisat2 on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 20, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - C.virginica Gonad RNAseq with FastP on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSummary - Geoduck RNAseq Data\n\n\n\n\n\n\n\n2021\n\n\nProject Summary\n\n\n\n\n\n\n\n\n\n\n\nJul 12, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Analysis - Identification of Potential Contaminating Sequences in Panopea-generosa-v1.0 Assembly Using BlobToolKit on Mox\n\n\n\n\n\n\n\n2021\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 12, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQC - Yaamini’s C.virginica RNAseq and WGBS from ZymoResearch on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 29, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - S.salar Gene Annotations from NCBI RefSeq GCF_000233375.1_ICSASG_v2_genomic.gff for Shelly\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Yaamini’s C.virginica WGBS and RNAseq Data from ZymoResearch\n\n\n\n\n\n\n\n2021\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nMay 28, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Comparisons - Ostrea lurida Non-scaffold Genome Assembly Comparisons Using Quast on Swoose\n\n\n\n\n\n\n\n2021\n\n\nOlympia Oyster Genome Assembly\n\n\n\n\n\n\n\n\n\n\n\nMay 21, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Assessment - Olurida_v090 Using Quast on Swoose\n\n\n\n\n\n\n\n2021\n\n\nOlympia Oyster Genome Assembly\n\n\n\n\n\n\n\n\n\n\n\nMay 21, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olurida_v090 with BGI Illumina and PacBio Hybrid Using Wengan on Mox\n\n\n\n\n\n\n\n2021\n\n\nOlympia Oyster Genome Assembly\n\n\n\n\n\n\n\n\n\n\n\nMay 20, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - O.lurida BGI FastQs with FastP on Mox\n\n\n\n\n\n\n\n2021\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 18, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Submission - Validation of Olurida_v081.fa and Annotated GFFs Prior to Submission to NCBI\n\n\n\n\n\n\n\n2021\n\n\nOlympia Oyster Genome Assembly\n\n\n\n\n\n\n\n\n\n\n\nMay 13, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeatMasker - C.gigas Rosling NCBI Genome GCA_902806645.1 on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 4, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSingularity - RStudio Server Container on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 23, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Mapping - 10x-Genomics Trimmed FastQ Mapped to P.generosa v1.0 Assembly Using Minimap2 for BlobToolKit on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - P.generosa v1.0 Assembly Using DIAMOND BLASTx for BlobToolKit on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - P.generosa v1.0 Assembly Using BLASTn for BlobToolKit on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming P.generosa 10x Genomics HiC FastQs with fastp on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 1, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate on C.bairdi Transcriptome v4.0 on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - DIAMOND BLASTx on C.bairdi Transcriptome v4.0 on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - C.bairdi Transcriptome v4.0 on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 17, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v4.0 on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 17, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi Transcriptome v4.0 Using Trinity on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 17, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Extractions - C.bairdi RNAseq Reads from C.opilio BLASTx Matches with seqkit on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 16, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDIAMOND BLASTx - C.bairdi RNAseq vs C.opilio Genome Proteins on Mox\n\n\n\n\n\n\n\n2021\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 12, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate Hematodinium v1.7 on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 9, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate Hematodinium v1.6 on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 9, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Hematodinium Transcriptomes v1.6 and v1.7 with Trinity on Mox\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 8, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Gene ID Extraction from P.generosa Genome GFF Using Methylation Machinery Gene IDs\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 26, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Gene ID Extraction from P.generosa Genome GFF Using Methylation Machinery List\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 26, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - A.elegantissima ONT Fast5 from Jay Dimond\n\n\n\n\n\n\n\n2021\n\n\nSRA Submission\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Anthopleura elegantissima - aggregating anenome - NanoPore Genome Sequence from Jay Dimond\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 1, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - M.magister MBD-BSseq Libraries to Univ. of Oregon GC3F\n\n\n\n\n\n\n\n2021\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2021\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Comparisons - C.bairdi Transcriptomes Evaluations with DETONATE rsem-eval on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 29, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAlignments - C.bairdi RNAseq Transcriptome Alignments Using Bowtie2 on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 24, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Cockle Clam Gonad H and E Slides\n\n\n\n\n\n\n\n2020\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQC - M.magister MBD-BSseq Pool Test MiSeq Run on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - M.magister MBD-BSseq Pool Test MiSeq Run\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAlignment - C.gigas RNAseq to GCF_000297895.1_oyster_v9 Genome Using STAR on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Haws Lab C.gigas Ploidy pH WGBS\n\n\n\n\n\n\n\n2020\n\n\nSRA Submission\n\n\n\n\n\n\n\n\n\n\n\nDec 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - Haws Lab C.gigas Ploidy pH WGBS 10bp 5 and 3 Prime Ends Using fastp and MultiQC on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 6, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQc - C.gigas Ploidy pH WGBS Raw Sequence Data from Haws Lab on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 6, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.gigas Diploid-Triploid pH Treatments Ctenidia WGBS from ZymoResearch\n\n\n\n\n\n\n\n2020\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nDec 5, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - Ronits C.gigas Ploidy WGBS 10bp 5 and 3 Prime Ends Using fastp and MultiQC on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - M.magister MBD BSseq Libraries for MiSeq at NOAA\n\n\n\n\n\n\n\n2020\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Quantification - M.magister MBD BSseq Libraries with Qubit\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Cockle Clam Gonad Histology Cassettes for H and E\n\n\n\n\n\n\n\n2020\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - Ronits C.gigas Ploidy WGBS Using fastp and MultiQC on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - M.magister MBD BSseq Libraries\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD BSseq Library Prep - M.magister MBD-selected DNA Using Pico Methyl-Seq Kit\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - P.generosa Hemocytes from Shelly\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Ronits C.gigas Ploidy WGBS\n\n\n\n\n\n\n\n2020\n\n\nSRA Submission\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQc - C.gigas Ploidy WGBS Raw Sequence Data from Ronits Project on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - Crustacean Transcripome Completeness Evaluation Using BUSCO on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.gigas Ploidy WGBS from Ronits Project via ZymoResearch\n\n\n\n\n\n\n\n2020\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nNov 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - MultiQC on S.salar RNAseq from fastp and HISAT2 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 6, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHard Drive Upgrade - Gannet Synology Server\n\n\n\n\n\n\n\n2020\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nNov 5, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Alignments - S.salar HISAT2 BAMs to GCF_000233375.1_ICSASG_v2_genomic.gtf Transcriptome Using StringTie on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 4, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Alignments - Trimmed S.salar RNAseq to GCF_000233375.1_ICSASG_v2_genomic.fa Using Hisat2 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 3, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Selection - M.magister Sheared Gill gDNA 16 of 24 Samples Set 3 of 3\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 3, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Selection - M.magister Sheared Gill gDNA 8 of 24 Samples Set 2 of 3\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming - Shelly S.salar RNAseq Using fastp and MultiQC on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 29, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Selection - M.magister Sheared Gill gDNA 8 of 24 Samples Set 1 of 3\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing - M.magister gDNA Additional Shearing CH05-01_21 CH07-11 and Bioanalyzer\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing - M.magister gDNA Shearing All Samples and Bioanalyzer\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing - M.magister CH05-21 gDNA Full Shearing Test and Bioanalyzer\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing - M.magister gDNA Shear Testing and Bioanalyzer\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 21, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Mapping - C.bairdi 201002558-2729-Q7 and 6129-403-26-Q7 Taxa-Specific NanoPore Reads to cbai_genome_v1.01.fasta Using Minimap2 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.bairdi NanoPore Reads Extractions With Seqtk on Mephisto\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 13, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoPore Reads Extractions - C.bairdi Taxonomic Reads Extractions with MEGAN6 on 201002558-2729-Q7 and 6129-403-26-Q7\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComparison - C.bairdi 20102558-2729 vs. 6129-403-26 NanoPore Taxonomic Assignments Using MEGAN6\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTaxonomic Assignments - C.bairdi 6129-403-26-Q7 NanoPore Reads Using DIAMOND BLASTx on Mox and MEGAN6 daa2rma on emu\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTaxonomic Assignments - C.bairdi 20102558-2729-Q7 NanoPore Reads Using DIAMOND BLASTx on Mox and MEGAN6 daa2rma on emu\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.bairdi NanoPore 6129-403-26 Quality Filtering Using NanoFilt on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.bairdi NanoPore 20102558-2729 Quality Filtering Using NanoFilt on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Assessment - BUSCO C.bairdi Genome v1.01 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 24, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Subsetting cbai_genome_v1.0 Assembly with faidx\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submissions - NanoPore C.bairdi 20102558-2729 and 6129_403_26\n\n\n\n\n\n\n\n2020\n\n\nSRA Submission\n\n\n\n\n\n\n\n\n\n\n\nSep 21, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Assessment - BUSCO C.bairdi Genome v1.0 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - C.bairdi NanoPore Quality Filtering Using NanoFilt on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - C.bairdi - cbai_v1.0 - Using All NanoPore Data With Flye on Mox\n\n\n\n\n\n\n\n2020\n\n\nGenome Assembly\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTaxonomic Assignments - C.bairdi NanoPore Reads Using DIAMOND BLASTx on Mox and MEGAN6 daa2rma on swoose\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Geoduck Normalizing Gene Primers 28s-v4 and EF1a-v1 Tests\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 16, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Geoduck Normalizing Gene Primer Checks\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Visualization of C.bairdi NanoPore Sequencing Using NanoPlot on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - NanoPore Fast5 Conversion to FastQ of C.bairdi 6129_403_26 on Mox with GPU Node\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 4, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - NanoPore Fast5 Conversion to FastQ of C.bairdi 20102558-2729 Run-02 on Mox with GPU Node\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 4, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - NanoPore Fast5 Conversion to FastQ of C.bairdi 20102558-2729 Run-01 on Mox with GPU Node\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 4, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submission - Supplemental Ronits C.gigas Diploid-Triploid Ctendidia gDNA for WGBS by ZymoResearch\n\n\n\n\n\n\n\n2020\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nSep 1, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Re-quant Ronits C.gigas Diploid-Triploid Ctenidia gDNA Submitted to ZymoResearch\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 1, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi v2.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nAug 31, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi v3.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nAug 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate Hematodinium v3.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate Hematodinium v2.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - C.bairdi Transcriptomes v2.1 and v3.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nAug 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - P.generosa RPL5 and TIF3s6b v2 and v3 Normalizing Gene Assessment\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 25, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submitted - C.gigas Diploid-Triploid pH Treatments Ctenidia to ZymoResearch for WGBS\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 24, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - P.generosa RPL5-v2-v3 and TIF3s6b-v2-v3 Primer Tests\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 24, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - C.gigas High-Low pH Triploid and Diploid Ctenidia\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 21, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - C.gigas High-Low pH Triploid Diploid from Maria Haws Lab\n\n\n\n\n\n\n\n2020\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nAug 20, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Ronits C.gigas Diploid and Triploid Ctenidia to ZymoResearch for WGBS\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 20, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Stats - C.bairdi Transcriptomes v2.1 and v3.1 Trinity Stats on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming-FastQC-MultiQC - Robertos C.gigas WGBS FastQ Data with fastp FastQC and MultiQC on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - Hematodinium Transcriptomes v1.6, v1.7, v2.1 and v3.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Stats - cbaiodinium Transcriptomes v2.1 and v3.1 Trinity Stats on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on Hematodinium v1.6 v1.7 v2.1 and v3.1 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Hematodinium Transcriptomes v1.6 v1.7 v2.1 v3.1 with DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - P.generosa APLP and TIF3s8-1 with cDNA\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQ Read Alignment and Quantification - P.generosa Water Metagenomic Libraries to MetaGeneMark Assembly with Hisat2 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPrimer Design and In-Silico Testing - Geoduck Reproduction Primers\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 30, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Testing P.generosa Reproduction-related Primers\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 29, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - P.generosa Metagenomics Data\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Testing P.generosa Reproduction-related Primers\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - C.gigas Diploid (Ronit) and Triploid (Nisbet)\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Geoduck Epigenetic Ocean Acidification RNAseq\n\n\n\n\n\n\n\n2020\n\n\nSRA Submission\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nENA Submission - Ostrea lurida draft genome Olurida_v081.fa\n\n\n\n\n\n\n\n2020\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Data Extractions Using MEGAN6\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Transcriptomes v2.1 and v3.1 Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v3.1\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v2.1\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequence Extractions - C.bairdi Transcriptomes v2.0 and v3.0 Excluding Alveolata with MEGAN6 on Swoose\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Transcriptomes v2.0 and v3.0 with DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 4, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Comparison - C.bairdi Transcriptomes Evaluations with DETONATE on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Comparison - C.bairdi Transcriptomes Compared with DETONATE on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 29, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi Transcriptome-v1.7 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 29, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Comparisons - C.bairdi BUSCO Scores\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - C.bairdi Transcriptome v1.7 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Transcriptome v1.7 Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v1.7\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi All Pooled Arthropoda-only RNAseq Data with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi Transcriptome-v3.0 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - P.trituberculatus (Japanese blue crab) NCBI SRA BioProject PRJNA597187 Data with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Library Assessment - Determine RNAseq Library Strandedness from P.trituberculatus SRA BioProject PRJNA597187\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 21, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTutorial - SRA Toolkit for Data Retrieval and Conversion to FastQ\n\n\n\n\n\n\n\n2020\n\n\nTutorials\n\n\n\n\n\n\n\n\n\n\n\nMay 21, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi Transcriptome-v1.6 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 20, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - C.bairdi Transcriptome v1.6 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - C.bairdi Transcriptome v3.0 from 20200518 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Transcriptome v1.6 Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v1.6\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Transcriptome v3.0 Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v3.0\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi All Arthropoda-specific RNAseq Data with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Arthropoda and Alveolata D26 Pool RNAseq FastQ Extractions\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi All Pooled RNAseq Data Without Taxonomic Filters with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi Transcriptome v2.0 from 20200502 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 13, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransDecoder - C.bairdi Transcriptome v2.0 from 20200502 on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Transcriptome v2.0 Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi v2.0 Transcriptome\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi All RNAseq Data Without Taxonomic Filters with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Abundance - C.bairdi Alignment-free with Salmon Using 2020-GW Data on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 29, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGO to GOslim - C.bairdi Enriched GO Terms from 20200422 DEGs\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 27, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQC - Laura Spencer’s QuantSeq Data\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGene Expression - C.bairdi Pairwise DEG Comparisons with 2019 RNAseq using Trinity-Salmon-EdgeR on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Reads Extractions - C.bairdi Taxonomic Reads Extractions with MEGAN6 on swoose\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 19, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscript Abundance - C.bairdi Alignment-free with Salmon on Mox for Grace\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - C.bairdi RNAseq Data\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimmingFastQCMultiQC—C.bairdi-RNAseq-FastQ-with-fastp-on-Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTaxonomic Assignments - C.bairdi RNAseq Using DIAMOND BLASTx on Mox and MEGAN6 Meganizer on swoose\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQC - C.bairdi Raw RNAseq from NWGSC\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Arthropoda and Alveolata Day and Treatment Taxonomic RNAseq FastQ Extractions\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.bairdi RNAseq from NWGSC\n\n\n\n\n\n\n\n2020\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi MEGAN6 Taxonomic-specific Trinity Assembly on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi MEGAN Trinity Assembly Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate Hematodinium MEGAN6 Taxonomic-specific Trinity Assembly on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransdecoder - Hematodinium MEGAN6 Taxonomic-Specific Reads Assembly from 20200330\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransdecoder - C.bairdi MEGAN6 Taxonomic-Specific Reads Assembly from 20200330\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi MEGAN Transcriptome\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Hematodinium MEGAN Trinity Assembly Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 31, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on Hematodinium MEGAN Transcriptome\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 31, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Hematodinium with MEGAN6 Taxonomy-specific Reads with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 30, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi with MEGAN6 Taxonomy-specific Reads with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 30, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Reads Extractions - C.bairdi Taxonomic Reads Extractions with MEGAN6 on swoose\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 30, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Using DIAMOND BLASTx on Mox and MEGAN6 Meganizer on swoose\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming/FastQC/MultiQC - C.bairdi RNAseq FastQ with fastp on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.bairdi RNAseq Data from Genewiz\n\n\n\n\n\n\n\n2020\n\n\nData Received\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - C.bairdi Hemocyte Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 11, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoPore Sequencing - C.bairdi gDNA 6129_403_26\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 11, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.bairdi RNA Check for Residual gDNA\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming/MultiQC - Methcompare Bisulfite FastQs with fastp on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 6, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - C.bairdi RNA from Hemolymph Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMar 6, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Create Canonical Olurida_v081 Genes FastA\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.bairdi RNA Check for Residual gDNA\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.bairdi Primer Tests on gDNA\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 25, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPrimer Design - C.bairdi Primers for Checking RNA for Residual gDNA\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 20, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - C.bairdi RNA from Samples 6212_132_9 6212_334_12 6212_485_26\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 11, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation & Quantification - C.bairdi RNA from Samples 6212_132_9 6212_334_12 6212_485_26\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 11, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation & Quantification - C.bairdi RNA from Sample 6129_403_26\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - Additional C.bairdi gDNA from Sample 6129_403_26\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation, Quantification, and Gel - C.bairdi gDNA Sample 6129_403_26\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on Hematodinium MEGAN Transcriptome\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assessment - BUSCO Metazoa on C.bairdi MEGAN Transcriptome\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGene Expression - Hematodinium MEGAN6 with Trinity and EdgeR\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGene Expression - C.bairdi MEGAN6 with Trinity and EdgeR\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Arthropoda and Alveolata Day and Treatment Taxonomic RNAseq FastQ Extractions\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 31, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - C.bairdi Hemocyte Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 28, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate Hematodinium MEGAN6 Taxonomic-specific Trinity Assembly on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Trinotate C.bairdi MEGAN6 Taxonomic-specific Trinity Assembly on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - C.bairdi Hemocyte Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 26, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - C.bairdi Hemocyte Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 24, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransdecoder - Hematodinium MEGAN6 Taxonomic-Specific Reads Assembly from 20200122\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransdecoder - C.bairdi MEGAN6 Taxonomic-Specific Reads Assembly from 20200122\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Hematodinium MEGAN Trinity Assembly Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi MEGAN Trinity Assembly Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - C.bairdi Hemocyte Pellets in RNAlater Troubleshooting\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quality Assessment - Agarose Gel for C.bairdi 20102558-2729 gDNA from 20200122\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Arthropoda and Alveolata Taxonomic RNAseq FastQ Extractions\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.bairdi 20102558-2729 EtOH-preserved Tissue via Three Variations Using Quick DNA-RNA MicroPrep Kit\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Hematodinium with MEGAN6 Taxonomy-specific Reads with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi with MEGAN6 Taxonomy-specific Reads with Trinity on Mox\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - C.bairdi Hemolymph Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - C.bairdi Hemolymph Pellets in RNAlater\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 17, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Reads Extractions - C.bairdi Taxonomic Reads Extractions with MEGAN6 on swoose\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLab Maintenance - Cluster UPS Battery Replacement\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 10, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoPore Sequencing - C.bairdi gDNA Sample 20102558-2729\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quality Assessment - Agarose Gel and NanoDrop on C.bairdi gDNA\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - C.bairdi gDNA from EtOH Preserved Tissue\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoPore Sequencing - Initial NanoPore MinION Lambda Sequencing Test\n\n\n\n\n\n\n\n2020\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 7, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Using DIAMOND BLASTx on Mox and MEGAN6 Meganizer\n\n\n\n\n\n\n\n2020\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJan 3, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReagent Prep - RNA Pico Ladder Aliquoting and Testing\n\n\n\n\n\n\n\n2020\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nJan 2, 2020\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Trinity Assembly Trinotate on Mox\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - C.bairdi Trinity Assembly BLASTx on Mox\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 24, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransdecoder - C.bairdi De Novo Transcriptome from 20191218 on Mox\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 20, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - C.bairdi Trimmed RNAseq Using Trinity on Mox\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming/FastQC/MultiQC - C.bairdi RNAseq FastQ with fastp on Mox\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Olurida_v081 UTR GFFs and Intergenic, Intron BED files\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 14, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Pacific Oyster Tissues from Hawaii (Maria Haws) from High and Low pCO2\n\n\n\n\n\n\n\n2019\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nDec 10, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Renaming, Splitting, and Feature Counts of Updated Pgenerosa_v074 GenSAS Merged GFF\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 3, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Crassostrea gigas and sikamea Mantle gDNA from Marinelli Shellfish Company\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 3, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - C.bairdi Hemolymph and Tissue in Ethanol from Pam Jensen\n\n\n\n\n\n\n\n2019\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nNov 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Geoduck hemolymph and hemocyte cDNA with vitellogenin primers\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - P.generosa DNased Hemolypmh and Hemocyte RNA from 20191125\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - Geoduck hemolymph and hemocyte samples\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Crassostrea gigas and sikamea Mantle gDNA from Marinellie Shellfish Company - No Multiplex\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Crassostrea gigas and sikamea Mantle gDNA from Marinelli Shellfish Company\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Rename Pgenerosa_v074 Bismark Coverage Files Scaffold Names\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 20, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Crassostrea gigas and sikamea Mantle gDNA from Marinelli Shellfish Company\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 19, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - Crassostrea gigas and Crassostrea sikamea Mantle Tissue from Marinelli Shellfish Company\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 19, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Additional Features Stats for Panopea-generosa-v1.0\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 6, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Rename Pgenerosa_v074 Files and Scaffolds\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 5, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Splitting BAM by Size for Upload to OSF\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 31, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Marinelli Shellfish Company C.gigas and C.sikamea Oysters\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 30, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Create Panopea-generosa-vv0.74.a4 Intron and Intergenic BED Files\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 30, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Feature Counts - Panopea-generosa-vv0.74.a4\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 29, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQC - C.bairdi RNAseq Day 12 26 Infected Uninfected\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nOct 24, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.bairdi RNAseq Day9-12-26 Infected-Uninfected\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\nData Received\n\n\n\n\n\n\n\n\n\n\n\nOct 24, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLab Maintenance - Cluster UPS Battery Replacement\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 22, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics Annotation - P.generosa Water Samples with MEGAN6\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 14, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.bairdi RNAseq Day9-12-26 Infected-Uninfected\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nOct 3, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v074 a4 Using GenSAS\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics Annotation - P.generosa Water Samples Using DIAMOND BLASTx on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimming/FastQC/MultiQC - P.generosa EPI FastQs with FASTP on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Hollie’s Juvenile OA BS-seq Data\n\n\n\n\n\n\n\n2019\n\n\nSRA Submissions\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Pgenerosa_v074.a3 Annotation Genome Repeats Compostion\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 5, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Pgenerosa_v074.a3 Annotation Genome Feature Sequence Lengths\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 4, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Panopea generosa Genome Feature Sequence Lengths\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Create a CpG GFF from Pgenerosa_v074 using EMBOSS fuzznuc on Swoose\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v074 MAKER on Mox with Stringtie Transcripts GFF\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 19, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs Pgenerosa_v070 with MUMmer Promer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs S.glomerata NCBI with MUMmer Promer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs M.yessoensis NCBI with MUMmer Promer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs H.sapiens NCBI with MUMmer Promer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs C.virginica NCBI with MUMmer Promer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs C.gigas NCBI with MUMmer Promer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs Pgenerosa_v070 with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs Pgenerosa_v074 with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs S.glomerata NCBI with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs M.yessoensis NCBI with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs C.gigas NCBI with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs H.sapiens NCBI with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Comparison - Pgenerosa_v074 vs C.virginica NCBI with MUMmer on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeatMasker - Pgenerosa_v070 for Transposable Element ID on Roadrunner\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - FastA Splitting With faSplit\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Summary - P.generosa Transcriptome Assemblies Stats\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 29, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Compression - P.generosa Transcriptome Assemblies Using CD-Hit-est on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 29, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v074 Transcript Isoform ID with Stringtie on Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v070 Transcript Isoform ID with Stringtie on Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v074 Hisat2 Transcript Isoform Index\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v070 Hisat2 Transcript Isoform Index\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v070 and v074 Top 18 Scaffolds Feature Count Comparisons\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 22, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - O.lurida 20190709-v081 Transcript Isoform ID with Stringtie on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 16, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - O.lurida 20190709-v081 Hisat2 Transcript Isoform Index\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 16, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assessment - BUSCO Metazoa on Pgenerosa_v074 on Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assessment - BUSCO Metazoa on Pgenerosa_v70 on Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v071 Using GenSAS\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v074 Using GenSAS\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Olurida_v081 with MAKER and Tissue-specific Transcriptomes on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v074 MAKER on Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 1, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Juvenile Ambient OA EPI123 with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Juvenile Super Low OA EPI115 with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Larvae Day5 EPI99 with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Gonad with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Juvenile Ambient OA EPI124 with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Ctenidia with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Super Low OA EPI116 with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeatModeler - Pgenerosa_v074 for MAKER Annotation on Emu\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeatMasker - Pgenerosa_v074 for Transposable Element ID on Roadrunner\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - C.virginica Mantle MBD-BSseq from ZymoResearch\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - FastA Subsetting of Pgenerosa_v070.fa Using samtools faidx\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - O.lurida (v081) Transcript Isoform ID with Stringtie on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - O.lurida (v081) Hisat2 Transcript Isoforms Index\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Refining Anvio Binning\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 19, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Taxonomic Diversity and Sequencing Coverage with MEGAHIT BLASTx and Krona Plots\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 12, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - Create Pgenerosa_v070 GFFs\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Tanner Crab Infected vs Uninfected RNAseq\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nMay 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - BLASTx of Individual Water Sample MEGAHIT Assemblies on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Decisions - C.bairdi RNAs for Library Pools\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC-MultiQC - Additional C.gigas WGBS Sequencing Data from Genewiz Received 20190501\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 1, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Additional C.gigas Whole Genome Bisulfite Sequencing Data from Genewiz\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 1, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - C.bairdi Hemolymph Pellet in RNAlater\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 30, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - P.generosa 10x Genomics Sequencing Data\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 24, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - C.virginica MBD Sequencing Coverage\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics Annotation - P.generosa Water Samples Using BLASTn on Mox and KronaTools Visualization to Compare pH Treatments\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 16, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics Gene Prediction - P.generosa Water Samples Using MetaGeneMark on Mox to Compare pH Treatments\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 16, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC - WGBS Sequencing Data from Genewiz Received 20190408\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenome Assemblies - P.generosa Water Samples Trimmed HiSeqX Data Using Megahit on Mox to Compare pH Treatments\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation and Quantification - Crab Hemolypmh Using Quick-DNA-RNA Microprep Plus Kit\n\n\n\n\n\n\n\n2019\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nApr 11, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Larvae Day5 EPI99 with HiSeq and NovaSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Gonad HiSeq and NovaSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Juvenile Ambient OA EPI124 with HiSeq and NovaSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Juvenile Ambient OA EPI123 with HiSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Juvenile Super Low OA EPI116 with HiSeq and NovaSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Juvenile Super Low OA EPI115 with HiSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Ctenidia with HiSeq and NovaSeq Data on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Whole Genome Bisulfite Sequencing Data from Genewiz Received\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - P.generosa Water Sample Assembly Comparisons with Quast\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Geoduck Water Sample Assembly Comparisons with MetaQuast\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 3, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Taxonomic Diversity Comparisons from Geoduck Water with Anvio on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 1, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenome Assemblies - P.generosa Water Samples Trimmed HiSeqX Data Using Megahit on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransposable Element Mapping - Crassostrea gigas Genome v9 Using RepeatMasker 4.07 on Roadrunner\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 27, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Lotterhos C.virginica Mantle MBD DNA to ZymoResearch for BSseq\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - SRA Submission Panopea generosa Day 5 Larvae RNAseq\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Taxonomic Diversity from Geoduck Water with BLASTn and Krona Plots\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenomics - Taxonomic Diversity from Geoduck Water with BLASTp and Krona plots\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 25, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment - DNA Quantification of C.virginica MBD Samples from 20190312\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 19, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Juvenile Day 5 with Trinotate on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Heart with Trinotate on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Ctenidia with Trinotate on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Gonad with Trinotate on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Heart with BLASTx on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Gonad with BLASTx on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Ctenidia with BLASTx on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Juvenile Day 5 with BLASTx on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Juvenile Day 5 with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Heart with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Gonad with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Annotation - Geoduck Ctenidia with Transdecoder on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMethylation Analysis - C.virginica Gonad MBD with Varying Read Subsets with Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Create C.virginica Bisulfite Genome with Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 12, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment - Ethanol Precipitation of C.virginica MBD samples\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 12, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment - C.virginica Sheared Mantle DNA from 20190306\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 11, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment - C.virginica Sheared Mantle DNA from 20190306\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing & Bioanalyzer - Lotterhos C.virginica Mantle gDNA from 2018114\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 6, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Lotterhos C.virginica Mantle DNA from 20181114\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 5, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Data Migration and Drive Expansion on Gannet\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 4, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assessment - BUSCO Metazoa on P.generosa v071 on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v070 MAKER on Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Wrangling - CpG OE Calculations on C.virginica Genes\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 26, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMethylation Analysis - O.lurida Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 22, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMethylation Analysis - P.generosa Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 22, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMethylation Analysis - C.virginica\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 22, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMethylation Analysis - C.gigas Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 22, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Create C.virginica Bisulfite Genome wit Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Create C.gigas Bisulfite Genome with Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Create Geoduck Bisulfite Genomes with Bismark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 21, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Juvenile Day 5\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Heart\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Gonad\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck Tissue-specific Assembly Ctenidia\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Pgenerosa_v71 with MAKER on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 13, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequence Subsetting - Pgenerosa_v70 Genome Assembly with faidx\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 11, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Robertos C.gigas DNA for Whole Genome Bisulfite Sequencing (Genewiz)\n\n\n\n\n\n\n\n2019\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - C.virginica DNA and Tissues from Lotterhos Lab\n\n\n\n\n\n\n\n2019\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nJan 29, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - Ronit’s C.gigas Ploidy Ctenidia\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas Ploidy Experiment Ctenidia\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 17, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER BUSCO Metazoa Augustus Training on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Geoduck Genome with MAKER Submitted to Mox\n\n\n\n\n\n\n\n2019\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 15, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER BUSCO (eukaryota_odb9) Augustus Training Submitted to Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER Functional Annotations on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER Proteins BLASTp on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER Proteins InterProScan5 on Mox\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER ID Mapping\n\n\n\n\n\n\n\n2019\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGene Prediction - HiSeqX Metagenomics from Geoduck Water Using MetaGeneMark on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 3, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMetagenome Assembly - P.generosa Water Sample Trimmed HiSeqX Data Using Megahit on Mox\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 2, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nVCF Splitting - C.virginica VCF Using BCFtools\n\n\n\n\n\n\n\n2019\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 2, 2019\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas Ploidy Experiment Ctenidia\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 20, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeat Library Construction - P.generosa RepeatModeler v1.0.11\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Relative mitochondrial abundance in C.gigas diploids and triploids subjected to acute heat stress via COX1\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBLASTx - Clupea pallasii (Pacific herring) liver and testes transcriptomes on Mox\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 12, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC and Trimming - Metagenomics (Geoduck) HiSeqX Reads from 20180809\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPrimer Design - Gigas COX1 using Primer3\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Geoduck gonad cDNA with vitellogenin primers\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 6, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Geoduck gonad RNA pool\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 6, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPrimer Design - Geoduck Vitellogenin using Primer3\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 29, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Olurida_v081 MAKER on Mox\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 27, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAnnotation - Geoduck Transcritpome with TransDecoder\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 21, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Quantification - Ronit’s C.gigas DNased RNA from 20181115\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 20, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Ronit’s C.gigas DNased RNA from 20181115\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 20, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Ronit’s DNased C.gigas RNA with Elongation Factor Primers\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 15, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - Ronit’s C.gigas Ctenidia RNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 15, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation and Quantification - Lotterhos C.virginica Mantle DNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Install NCBI nr nt BLAST Database on Mox\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMy New Notebook\n\n\n\n\n\n\n\n2018\n\n\n\n\n\n\n\n\n\n\n\n\n\n\nNov 8, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRepeat Library Construction - O.lurida RepeatModeler v1.0.11\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 22, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - Ronit’s C.gigas ploidy/dessication/heat stress cDNA (1:5 dilution)\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 18, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Crassostrea virginica (Eastern oyster) tissue from Lotterhos Lab (Northeastern University)\n\n\n\n\n\n\n\n2018\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nOct 17, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Ronit’s C.gigas DNased ctenidia RNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 17, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Ronit’s DNAsed C.gigas Ploidy/Dessication RNA with elongation factor primers\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 17, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR – C.gigas primer and gDNA tests with 18s and EF1 primers\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Ronit’s DNAsed C.gigas Ploidy/Dessication RNA with 18s primers\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Ronit’s C.gigas Ploiyd/Dessication Ctenidia RNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Quantification - Ronit’s C.gigas Ploidy/Dessication RNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Chionoecetes bairdi RNAseq & FastQC Analysis\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 15, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nVCF Splitting with bcftools\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 15, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Tanner Crab Hemolymph Pellet in RNAlater using TriReagent\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Ronit’s C.gigas diploid/triploid dessication/heat shock ctenidia tissues\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 3, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Olymia oyster Whole Genome BS-seq Data\n\n\n\n\n\n\n\n2018\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 3, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nInstallation - Microsoft Machine Learning Server (Microsoft R Open) on Emu/Roadrunner R Studio Server\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nUbuntu – Fix “No Video Signal” Issue on Emu\n\n\n\n\n\n\n\n2018\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Alignment & Bedgraph – Olympia oyster transcriptome with Olurida_v080 genome assembly\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 26, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBedgraph – Olympia oyster transcriptome with Olurida_v081 genome assembly\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 26, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Alignment – Olympia oyster RNAseq reads aligned to genome with HISAT2\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 25, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBedgraph - Olympia oyster transcriptome (FAIL)\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 24, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Alignment - Olympia oyster Trinity transcriptome aligned to genome with Bowtie2\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Olympia oyster RNAseq Data with Trinity\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Analysis – Olympia Oyster Whole Genome BSseq Bismark Pipeline MethylKit Comparison\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Lyophilized Tanner Crab Hemolymph in RNAlater\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Analysis - Olympia Oyster Whole Genome BSseq Bismark Pipeline Comparison\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 13, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Sea Lice DNA from 20180523\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 12, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC - C.virginica Oil Spill MBDseq Concatenated Sequences\n\n\n\n\n\n\n\n2018\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing Data Analysis - C.virginica Oil Spill MBDseq Concatenation & FastQC\n\n\n\n\n\n\n\n2018\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 10, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Analysis - Olympia oyster BSseq MethylKit Analysis\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 4, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTranscriptome Assembly - Geoduck RNAseq data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 4, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC/MultiQC/TrimGalore/MultiQC/FastQC/MultiQC - O.lurida WGBSseq for Methylation Analysis\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 30, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransposable Element Mapping – Crassostrea virginica Genome, Cvirginica_v300, using RepeatMasker 4.07\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 28, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Stats – Geoduck Hi-C Final Assembly Comparison\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 23, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Analysis - Bismark Pipeline on All Olympia oyster BSseq Datasets\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Geoduck Metagenome HiSeqX Data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation & Quantificaiton - Tanner Crab Hemolymph\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation – Olympia oyster genome annotation results #02\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 8, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Olympia oyster genome annotation results #01\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 8, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation – Olympia oyster genome complete - brief note\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Annotation - Olympia oyster genome using WQ-MAKER Instance on Jetstream\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 7, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - Tanner Crab RNA Isolated with RNeasy Plus Mini Kit\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 1, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Cleanup - Tanner Crab RNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Tanner Crab Hemolymph Using RNeasy Plus Mini Kit\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMox – Over quota: Olympia oyster genome annotation progress (using Maker 2.31.10)\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 30, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Cleanup - Tanner Crab RNA Pools\n\n\n\n\n\n\n\n2018\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nJul 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMox - Olympia oyster genome annotation progress (using Maker 2.31.10)\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMox - Password-less SSH!\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nUbuntu - Fix “No Video Signal” Issue on Emu/Roadrunner\n\n\n\n\n\n\n\n2018\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJul 5, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransposable Element Mapping – Olympia Oyster Genome Assembly, Olurida_v081, using RepeatMasker 4.07\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 3, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Construction - Geoduck Water Filter Metagenome with Nextera DNA Flex Kit (Illumina)\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 6, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBS-seq Mapping – Olympia oyster bisulfite sequencing: Bismark Continued\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 31, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransposable Element Mapping – Crassostrea virginica NCBI Genome Assembly using RepeatMasker 4.07\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 29, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Mapping – Olympia oyster 2bRAD Data with Bowtie2 (on Mox)\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 26, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTransposable Element Mapping - Olympia Oyster Genome Assembly using RepeatMasker 4.07\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Received - Sea lice DNA from Cris Gallardo-Escarate at Universidad de Concepción\n\n\n\n\n\n\n\n2018\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Installation – RepeatMasker v4.0.7 on Emu/Roadrunner Continued\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Installation - RepeatMasker v4.0.7 on Emu/Roadrunner\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 22, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC – TrimGalore! RRBS Geoduck BS-seq FASTQ data (directional)\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC - RRBS Geoduck BS-seq FASTQ data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC – TrimGalore! RRBS Geoduck BS-seq FASTQ data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Illumina NovaSeq Geoduck Genome Sequencing\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly - Geoduck Hi-C Assembly Subsetting\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 12, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRead Mapping - Mapping Illumina Data to Geoduck Genome Assemblies with Bowtie2\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBS-seq Mapping - Olympia oyster bisulfite sequencing: TrimGalore > FastQC > Bismark\n\n\n\n\n\n\n\n2018\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly & Stats - SparseAssembler (k95) on Geoduck Sequence Data > Quast for Stats\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 1, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Stats - Geoduck Genome Assembly Comparisons w/Quast - SparseAssembler, SuperNova, Hi-C\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 30, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Stats - Geoduck Hi-C Assembly Comparison\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 30, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - Metagenomics Water Filters\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTotal Alkalinity Calculations - Yaamini’s Ocean Chemistry Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 24, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly – SparseAssembler (k 111) on Geoduck Sequence Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 23, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly – SparseAssembler (k 131) on Geoduck Sequence Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 22, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Geoduck Phase Genomics Hi-C Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nApr 21, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nKmer Estimation – Kmergenie (k 301) on Geoduck Sequence Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 21, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nKmer Estimation – Kmergenie Tweaks on Geoduck Sequence Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nKmer Estimation - Kmergenie on Geoduck Sequence Data (default settings)\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Stats - Quast Stats for Geoduck SparseAssembler Job from 20180405\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - SRA Submission LSU C.virginica Oil Spill MBD BS-seq Data\n\n\n\n\n\n\n\n2018\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 12, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC - Trim 10bp 5’/3’ ends C.virginica MBD BS-seq FASTQ data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - Metagenomics Water Filters\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC - 2bp 3’ end Read 1s Trim C.virginica MBD BS-seq FASTQ data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC - 14bp Trim C.virginica MBD BS-seq FASTQ data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore/FastQC/MultiQC - Auto-trim C.virginica MBD BS-seq FASTQ data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC/MultiQC - C. virginica MBD BS-seq Data\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - SparseAssembler Geoduck Genomic Data, kmer=101\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 5, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGunzip - BGI HiSeq Geoduck Genome Sequencing Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 5, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGunzip - Trimmed Illumina Geoduck HiSeq Genome Sequencing Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 4, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification – Geoduck larvae metagenome filter rinses\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 3, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrations - Yaamini’s Seawater Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 3, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrations - Yaamini’s Seawater Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 2, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore!/FastQC/MultiQC - Illumina HiSeq Genome Sequencing Data Continued\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 1, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Crassostrea virginica MBD BS-seq from ZymoResearch\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 29, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC/MultiQC – Illumina HiSeq Genome Sequencing Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 28, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrimGalore!/FastQC/MultiQC - Illumina HiSeq Genome Sequencing Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 28, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFastQC/MultiQC - BGI Geoduck Genome Sequencing Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 27, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrations - Hollie’s Seawater Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 27, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly - Geoduck NovaSeq using SparseAssembler kmer = 101\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 27, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly - Geoduck NovaSeq using SparseAssembler (TL;DR - it worked!)\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 22, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrations - Hollie’s Seawater Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 20, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - Geoduck larvae metagenome filter rinses\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 20, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrations - Hollie’s Seawater Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrations - Hollie’s Seawater Samples\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Geoduck larvae metagenome filter rinses\n\n\n\n\n\n\n\n2018\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nMar 13, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly - Geoduck NovaSeq using SparseAssembler (failed)\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 8, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nProgress Report - Titrator\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 6, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nUbuntu Installation - Convert Apple Xserve “bigfish” to Ubuntu\n\n\n\n\n\n\n\n2018\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHardware Upgrades - USB 3.0 PCI Card and 1TB SSD in Woodpecker\n\n\n\n\n\n\n\n2018\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Triploid Crassostrea gigas from Nisbet Oyster Company\n\n\n\n\n\n\n\n2018\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNovaSeq Assembly - The Struggle is Real - Real Annoying!\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 21, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly - Geoduck Illumina NovaSeq SOAPdenovo2 on Mox (FAIL)\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 19, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation & DNA Quantification - C. virginica MBD DNA from Yaamini\n\n\n\n\n\n\n\n2018\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nFeb 7, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNovaSeq Assembly - Trimmed Geoduck NovaSeq with Meraculous\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 5, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTitrator Setup - Functional Methods & Data Exports\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 1, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAdapter Trimming and FASTQC - Illumina Geoduck Novaseq Data\n\n\n\n\n\n\n\n2018\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 25, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Install - 10x Genomics Supernova on Mox (Hyak)\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - C. virginica gDNA, MBD, and MspI to Qiagen\n\n\n\n\n\n\n\n2018\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nJan 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Comparisons – Oly Assemblies Using Quast\n\n\n\n\n\n\n\n2018\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 16, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - MspI-digested Crassostrea virginica gDNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPhenol:Chloroform Extractions and EtOH Precipitations - MspI Digestions of C.virginica DNA from Earlier Today\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digestion - MspI on Crassotrea virginica gDNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - C.virginica MBD-enriched DNA\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 10, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment – Crassostrea virginica Sheared DNA Day 3\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 10, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment – Crassostrea virginica Sheared DNA Day 2\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment - Crassostrea virginica Sheared DNA Day 1\n\n\n\n\n\n\n\n2018\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2018\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTissue Sampling - Crassostrea virginica Tissues for Archiving\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 13, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Install - MSMTP For Email Notices of Bash Job Completion on Emu (Ubuntu)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 13, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Pulverized Geoduck Tissues to Illumina for More 10x Genomics Sequencing\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 12, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Sonication & Bioanalzyer - C. virginica gDNA for MeDIP\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - Crassostrea virginica Mantle gDNA\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly – Olympia Oyster Illumina & PacBio Using PB Jelly w/BGI Scaffold Assembly\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTroubleshooting – PB Jelly Install on Emu Continued\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Geoduck Tissues to Illumina for More 10x Genomics Sequencing\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nNov 20, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTroubleshooting - PB Jelly Install on Emu\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 20, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation & Quantification - C. virginica Gonad gDNA\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Comparison - Oly Assemblies Using Quast\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia Oyster Illumina & PacBio Using PB Jelly w/BGI Scaffold Assembly\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia Oyster Illumina & PacBio Using PB Jelly w/Platanus Assembly\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Tanner Crab Hemolymph in RNA Later from Pam Jensen\n\n\n\n\n\n\n\n2017\n\n\nSamples Received\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation & Quantification - Tanner crab hemolymph\n\n\n\n\n\n\n\n2017\n\n\nTanner Crab RNAseq\n\n\n\n\n\n\n\n\n\n\n\nNov 7, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Installation - ALPACA on Roadrunner\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 31, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Crash - Olympia oyster genome assembly with Masurca on Mox\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 31, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Installation - PB Jelly Suite and Blasr on Emu\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 30, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster Illumina & PacBio Reads Using Redundans\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 24, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Comparison - Oly PacBio Canu: Sam vs. Sean with Quast\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 23, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFAIL - Missing Data on Owl!\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 23, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster Illumina & PacBio reads using MaSuRCA\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 19, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Installation - MaSuRCA v3.2.3 Assembler on Mox (Hyak)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 19, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFail - Directory Contents Deleted!\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 18, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster PacBio Canu v1.6\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 18, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Convert Oly PacBio H5 to FASTQ\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster Redundans/Canu vs. Redundans/Racon\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 6, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia Oyster Redundans with Illumina + PacBio\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - minimap/miniasm/racon Overview\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 4, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAssembly Comparisons - Olympia oyster genome assemblies\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 3, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - C.virginica gonad tissue from Katie Lotterhos\n\n\n\n\n\n\n\n2017\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nOct 3, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - October 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster PacBio minimap/miniasm/racon\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster PacBio minimap/miniasm/racon\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenome Assembly - Olympia oyster PacBio minimap/miniasm/racon\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 7, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Submitted - Geoduck Ctenidia to Illumina for 10x Genomics Sequencing\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nAug 28, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nProject Progress - Olympia Oyster Genome Assemblies by Sean Bennett\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 28, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Illumina Geoduck HiSeq & MiSeq Data\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 8, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - August 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nAug 3, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Geoduck Genome Sequencing by Illumina\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Olympia oyster gonad RNA to Katherine Silliman @ Univ. of Chicago\n\n\n\n\n\n\n\n2017\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nJul 20, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Olympia oyster gonad tissue in paraffin histology blocks\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 19, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Olympia oyster gonad tissue in paraffin histology blocks\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - SRA Submission Olympia Oyster UW PacBio Data from 20170323\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management – Tarball of Olympia oyster UW PacBio Data from 20170323\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - OLYMPIA OYSTER UW PACBIO DATA (FROM 20170323) TO NIGHTINGALES\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 5, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Annotation - Olympia oyster histology blocks (from Laura Spencer)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 5, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nManuscript Re-submission - Oly Stress Response to PeerJ for Review\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 3, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Olympia oyster UW PacBio Data from 20170323\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJul 3, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGitHub Curation\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 22, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - June 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Quantification - Acropora cervicornis (Staghorn coral) DNA from Javier Casariego (FIU)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 11, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Acropora cervicornis (Staghorn coral) DNA from Javier Casariego (FIU)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 11, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Concentration - Acropora cervicornis (Staghorn coral) DNA from Javier Casariego (FIU)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 10, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Acropora cervicornis (Staghorn coral) DNA from Javier Casariego (FIU)\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 10, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Olympia oyster Histology Blocks and Slides (for Laura Spencer)\n\n\n\n\n\n\n\n2017\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nMay 9, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Acropora cervicornis (Staghorn coral) DNA from Javier Casariego (FIU)\n\n\n\n\n\n\n\n2017\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nMay 9, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - May 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nMay 2, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nManuscript Writing - Submitted!\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nApr 19, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nManuscript - Oly GBS 14 Day Plan\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nApr 18, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Olympia oyster PacBio Data\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 23, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management – SRA Submission Oly GBS Batch Submission\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 21, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Owl Partially Restored\n\n\n\n\n\n\n\n2017\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nMar 21, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - SRA Submission Oly GBS Batch Submission Fail\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 20, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTroubleshooting - Synology NAS (Owl) Down After Update\n\n\n\n\n\n\n\n2017\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing – Oly BGI GBS Reproducibility; fail?\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 17, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing – Oly BGI GBS Reproducibility Fail (but, less so than last time)…\n\n\n\n\n\n\n\n2017\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Oly BGI GBS Reproducibility Fail\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 7, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFASTQC - Oly BGI GBS Raw Illumina Data Demultiplexed\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 7, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFASTQC - Oly BGI GBS Raw Illumina Data\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - March 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nManuscript Writing - More “Nuances” Using Authorea\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Jay’s Coral RADseq and Hollie’s Geoduck Epi-RADseq\n\n\n\n\n\n\n\n2017\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 27, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - SRA Submission of Ostrea lurida GBS FASTQ Files\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\nSRA Submissions\n\n\n\n\n\n\n\n\n\n\n\nFeb 8, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - February 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nFeb 2, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nManuscript Writing - The “Nuances” of Using Authorea\n\n\n\n\n\n\n\n2017\n\n\nGenotype-by-sequencing at BGI\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission – Geoduck gDNA for Illumina Pilot Sequencing Project\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nJan 5, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck gDNA for Illumina-initiated Sequencing Project\n\n\n\n\n\n\n\n2017\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 5, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Replacement of Corrupt BGI Oly Genome FASTQ Files\n\n\n\n\n\n\n\n2017\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 4, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - January 2017\n\n\n\n\n\n\n\n2017\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nJan 3, 2017\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Geoduck RRBS Data Integrity Verification\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 30, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Geoduck RRBS Sequencing Data\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 29, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Geoduck Tissue & gDNA for Illumina Pilot Sequencing Project\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 21, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck gDNA for Potential Illumina-initiated Sequencing Project\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 21, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Geoduck Reduced Representation Bisulfite Pooled Libraries\n\n\n\n\n\n\n\n2016\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 20, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Ostrea lurida gDNA for PacBio Sequencing\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 19, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Integrity Check of Final BGI Olympia Oyster & Geoduck Data\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 15, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Ostrea lurida DNA for PacBio Sequencing\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Managment - Trim Output Cells from Jupyter Notebook\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Download Final BGI Genome & Assembly Files\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - December 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Continued O.lurida Fst Analysis from GBS Data\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - An Excercise in Futility\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Initial O.lurida Fst Determination from GBS Data\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 17, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Tracking O.lurida FASTQ File Corruption\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 17, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputer Management - Additional Configurations for Reformatted Xserves\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nNov 16, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Modify Eagle/Owl Cloud Sync Account\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nNov 7, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Retrieve data from Amazon EC2 Instance\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 4, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - November 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nNov 2, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management – Geoduck Small Insert Library Genome Assembly from BGI\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 25, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - October 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nOct 3, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received – Jay’s Coral epiRADseq - Not Demultiplexed\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 19, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nOyster Sampling - Olympia Oyster OA Populations at Manchester\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 12, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Jay’s Coral epiRADseq\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 8, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - September 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nSep 6, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Synology Cloud Sync to UW Google Drive\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nAug 29, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nServer HDD Failure – Owl\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nAug 22, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nManuscript Submission - Oly Stress Response to PeerJ for Review\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 18, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - fastStructure Population Analysis of Oly GBS PyRAD Output\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nAug 16, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Amazon EC2 Cost “Analysis”\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 1, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - August 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nAug 1, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - PyRad Analysis of Olympia Oyster GBS Data\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nJul 27, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Not Enough Power!\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 18, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Amazon EC2 Instance Out of Space?\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nJul 17, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - A Very Quick “Guide” to Amazon EC2 Continued\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDissection - Frozen Geoduck & Pacific Oyster\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nFilter Replacement - Xserve Server Rack Enclosure\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - The Very Quick “Guide” to Amazon Web Services Cloud Computing Instances (EC2)\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 13, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Quantification - Coral DNA from Jose M. Eirin-Lopez (Florida International University)\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 7, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - July 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nJul 1, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Coral DNA from Jose M. Eirin-Lopez (Florida International University)\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Coral DNA from Jose M. Eirin-Lopez (Florida International University)\n\n\n\n\n\n\n\n2016\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nJun 15, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDocker - VirtualBox Defaults on OS X\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJun 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDocker - One liner to create Docker container\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 9, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRAM Upgrade - Roadrunner (Apple Xserve) to 48GB RAM\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJun 9, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - June 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nJun 6, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDocker - Improving Roberts Lab Reproducibility\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputer Setup - Cluster Node003 Conversion\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nMay 31, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Oly GBS Data Using Stacks 1.37\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Olympia Oyster Small Insert Library Genome Assembly from BGI\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGBS Frustrations\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 3, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - May 2016\n\n\n\n\n\n\n\n2016\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nMay 2, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Release - Transcriptomic Profiles of Adult Female & Male Gonads in Panopea generosa (Pacific geoduck)\n\n\n\n\n\n\n\n2016\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\nSRA Submissions\n\n\n\n\n\n\n\n\n\n\n\nApr 28, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - O.lurida Raw BGI GBS FASTQ Data\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nApr 27, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nComputing - Speed Benchmark Comparisons Between Local, External, & Server Files\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 27, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Subset Olympia Oyster GBS Data from BGI as Single Population Using PyRAD\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nApr 18, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - Concatenate FASTQ files from Oly MBDseq Project\n\n\n\n\n\n\n\n2016\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nApr 11, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Oly GBS Data from BGI Using Stacks\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nApr 6, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSoftware Install - samtools-0.1.19 and stacks-1.37\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nApr 6, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission – Genome sequencing of the Olympia oyster (Ostrea lurida)\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\nSRA Submissions\n\n\n\n\n\n\n\n\n\n\n\nMar 28, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission – Genome sequencing of the Pacific geoduck (Panopea generosa)\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\nSRA Submissions\n\n\n\n\n\n\n\n\n\n\n\nMar 28, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Transcriptomic Profiles of Adult Female & Male Gonads in Panopea generosa (Pacific geoduck).\n\n\n\n\n\n\n\n2016\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nMar 24, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSRA Submission - Individual Transcriptomic Profiles of C.gigas Before & After Heat Shock\n\n\n\n\n\n\n\n2016\n\n\nSRA Submissions\n\n\n\n\n\n\n\n\n\n\n\nMar 23, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - SRA Submission Overview\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 23, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - O. lurida genotype-by-sequencing (GBS) data from BGI\n\n\n\n\n\n\n\n2016\n\n\nGenotype-by-sequencing at BGI\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Initial Geoduck Genome Assembly from BGI\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Initial Olympia oyster Genome Assembly from BGI\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - O.lurida 2bRAD Dec2015 Undetermined FASTQ files\n\n\n\n\n\n\n\n2016\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nMar 8, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Management - High-throughput Sequencing Data\n\n\n\n\n\n\n\n2016\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 4, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Ostrea lurida MBD-enriched BS-seq\n\n\n\n\n\n\n\n2016\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nFeb 3, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Ostrea lurida genome sequencing files from BGI\n\n\n\n\n\n\n\n2016\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 27, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Panopea generosa genome sequencing files from BGI\n\n\n\n\n\n\n\n2016\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 27, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Identification of duplicate files on Eagle\n\n\n\n\n\n\n\n2016\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Bisulfite-treated Illumina Sequencing from Genewiz\n\n\n\n\n\n\n\n2016\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2016\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Received - Oly 2bRAD Illumina Sequencing from Genewiz\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 31, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - BS-seq Library Pool to Genewiz\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nIllumina Methylation Library Quantification - BS-seq Oly/C.gigas Libraries\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nIllumina Methylation Library Construction - Oly/C.gigas Bisulfite-treated DNA\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 21, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - Bisulfite-treated Oly/C.gigas DNA\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReagent Prep - RNA Pico 6000 Ladder\n\n\n\n\n\n\n\n2015\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisulfite Treatment - Oly Reciprocal Transplant DNA & C.gigas Lotterhos DNA for BS-seq\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Oly gDNA for BS-seq Libraries, Take Two\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Oly gDNA for BS-seq Libraries\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Oly gDNA for BS-seq\n\n\n\n\n\n\n\n2015\n\n\nBS-seq Libraries for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nDec 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - 2bRAD Libraries for Genewiz\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 10, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - C.gigas Tissue & DNA from Katie Lotterhos\n\n\n\n\n\n\n\n2015\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nDec 10, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Olympia oyster MBD-enriched DNA to ZymoResearch\n\n\n\n\n\n\n\n2015\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\nOlympia oyster reciprocal transplant\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 8, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Additional Olympia Oyster gDNA for Genome Sequencing @ BGI\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Additional Geoduck gDNA for Genome Sequencing @ BGI\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Storage - Synology DX513\n\n\n\n\n\n\n\n2015\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Oly Oyster Bay Tissues for GBS\n\n\n\n\n\n\n\n2015\n\n\nGenotype-by-sequencing at BGI\n\n\nOlympia oyster reciprocal transplant\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Oly Tissue & DNA from Katherine Silliman\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\nSamples Received\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quality Assessment - Geoduck & Olympia Oyster gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Olympia Oyster Outer Mantle gDNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck Ctenidia gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPhenol-Chloroform DNA Cleanup - Geoduck gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPhenol-Chloroform DNA Cleanup - Olympia Oyster gDNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck Adductor Muscle gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Olympia Oyster Outer Mantle gDNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - MBD-enriched Olympia oyster DNA\n\n\n\n\n\n\n\n2015\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 23, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation - Olympia oyster MBD\n\n\n\n\n\n\n\n2015\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBD Enrichment - Sonicated Olympia Oyster gDNA\n\n\n\n\n\n\n\n2015\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 21, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Sonication - Oly gDNA for MBD\n\n\n\n\n\n\n\n2015\n\n\nMBD Enrichment for Sequencing at ZymoResearch\n\n\n\n\n\n\n\n\n\n\n\nNov 19, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR – Oly RAD-Seq Library Quantification\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Oly RAD-Seq Library Quantification\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGel Extraction - Oly RAD-Seq Prep Scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quality Assessment - Geoduck, Oly & Oly 2SN\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR – Oly RAD-seq Prep Scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR – Oly RAD-seq Test-scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 12, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification & Quality Assessment - Oly 2SN gDNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 4, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification & Quality Assessment - Geoduck & Oly gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 4, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolations – Oly Fidalgo 2SN Ctenidia\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 3, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nOyster Sampling - Oly Fidalgo 2SN, 2HL, 2NF Reciprocal Transplants Final Samplings\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 30, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation – Geoduck & Olympia Oyster\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 29, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAdaptor Ligation – Oly AlfI-Digested gDNA for RAD-seq\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 29, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digest – Oly gDNA for RAD-seq w/AlfI\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 28, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTroubleshooting - Oly RAD-seq\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 23, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolations - Fidalgo 2SN Reciprocal Transplants Final Samplings\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 21, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Oly RAD-seq Prep Scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Oly RAD-seq Test-scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 12, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAdaptor Ligation – Oly AlfI-Digested gDNA for RAD-seq\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digest – Oly gDNA for RAD-seq w/AlfI\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Additional Geoduck gDNA for Genome Sequencing @ BGI\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Additional Olympia Oyster gDNA for Genome Sequencing @ BGI\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nOct 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Geoduck & Olympia Oyster gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Pooled geoduck gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Geoduck Adductor Muscle\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck & Olympia Oyster\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Oly RAD-seq Test-scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Oly RAD-seq Test-scale PCR\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nOct 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAdaptor Ligation - Oly AlfI-Digested gDNA for RAD-seq\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nOct 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - October 2015\n\n\n\n\n\n\n\n2015\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nOct 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digest - Oly gDNA for RAD-seq w/AlfI\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nSep 30, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Olympia Oyster gDNA for Genome Sequencing @ BGI\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nSep 21, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Geoduck gDNA for Genome Sequencing @ BGI\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 21, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nUninterruptible Power Supplies (UPS)\n\n\n\n\n\n\n\n2015\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Geoduck & Olympia oyster gDNA Integrity Check\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation - Geoduck & Olympia oyster gDNA\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Olympia oyster Whole Body gDNA Integrity Check\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Olympia oyster whole body\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nSep 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel – Geoduck gDNA Integrity Check\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation – Geoduck Adductor Muscle & Foot\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Geoduck & Olympia Oyster gDNA Integrity Check\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation – Olympia oyster adductor musle & mantle\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation – Geoduck Adductor Muscle & Foot\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Geoduck & Olympia Oyster gDNA Integrity Check\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation - Olympia oyster adductor musle & mantle\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation - Geoduck Adductor Muscle & Foot\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAgarose Gel - Geoduck & Olympia Oyster gDNA Integrity Check\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation - Olympia oyster adductor musle & mantle\n\n\n\n\n\n\n\n2015\n\n\nOlympia Oyster Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nServer Email Notifications Fix - Eagle\n\n\n\n\n\n\n\n2015\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nSep 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic DNA Isolation - Geoduck Adductor Muscle & Foot\n\n\n\n\n\n\n\n2015\n\n\nGeoduck Genome Sequencing\n\n\n\n\n\n\n\n\n\n\n\nAug 28, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRAD-Seq Library Prep Reagents\n\n\n\n\n\n\n\n2015\n\n\n2bRAD Library Tests for Sequencing at Genewiz\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nAug 19, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSsoFast EvaGreen Supermix Aliquots\n\n\n\n\n\n\n\n2015\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nAug 11, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription – O.lurida DNased RNA 1hr post-mechanical stress\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Jake’s O.lurida ctenidia 1hr post-mechanical stress DNased RNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Quantification - O.lurida 1hr post-mechanical heat stress DNased RNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - August 2015\n\n\n\n\n\n\n\n2015\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nAug 3, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission – Olympia oyster PCRs Sanger Sequencing\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nServer HDD Failure - Owl\n\n\n\n\n\n\n\n2015\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJul 31, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - O.lurida Ctenidia 1hr Post-Mechanical Stress RNA\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nJul 27, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation – O.lurida Ctenidia 1hr Post-Mechanical Stress\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nJul 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - O.lurida Ctenidia 1hr Post-Mechanical Stress\n\n\n\n\n\n\n\n2015\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSsoFast EvaGreen Supermix Aliquots\n\n\n\n\n\n\n\n2015\n\n\nReagent Prep\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAutomatic Notebook Backups - wget Script & Synology Task Scheduler\n\n\n\n\n\n\n\n2015\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Sea Pen luciferase\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGOALS - July 2015\n\n\n\n\n\n\n\n2015\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nOpticon2 Calibration\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNAseq Data Receipt - Geoduck Gonad RNA 100bp PE Illumina\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission – Olympia oyster PCRs Sanger Sequencing\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - O.lurida DNased RNA Controls and 1hr Heat Shock\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission – Olympia oyster & Sea Pen PCRs Sanger Sequencing\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGel Purification - Olympia Oyster and Sea Pen PCRs\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Olympia oyster PCRs Sanger Sequencing\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 12, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Geoduck Gonad for RNA-seq\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - June 2015\n\n\n\n\n\n\n\n2015\n\n\nGoals\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - Geoduck Gonad RNA Quality Assessment\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Subset of Jake’s O.lurida DNased RNA\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioinformatics – Trimmomatic/FASTQC on C.gigas Larvae OA NGS Data\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Re-run Jake’s O.lurida DNased RNA Samples NC1, SC1, SC2, SC4 from 20150514\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 21, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR – Jake’s O.lurida ctenidia DNased RNA (1hr Heat Shock Samples)\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR – Jake’s O.lurida ctenidia DNased RNA (Control Samples)\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Jake’s O.lurida Ctenidia RNA (1hr Heat Shock) from 20150506\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Jake’s O.lurida Ctenidia RNA (Controls) from 20150507\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nISO Creation - OpticonMonitor3 Disc Cloning\n\n\n\n\n\n\n\n2015\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Jake O.lurida ctenidia RNA (Heat Shock Samples) from 20150506\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 12, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Jake O.lurida ctenidia RNA (Control Samples) From 20150507\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 12, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation – Geoduck Gonad in Paraffin Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nMay 7, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation – Jake’s O. lurida Ctenidia Control from 20150422\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 7, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioinformatics - Trimmomatic/FASTQC on C.gigas Larvae OA NGS Data\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Jake’s O. lurida Ctenidia 1hr Heat Stress from 20150422\n\n\n\n\n\n\n\n2015\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation – Geoduck Gonad in Paraffin Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nMay 5, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBLAST - C.gigas Larvae OA Illumina Data Against GenBank nt DB\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nMay 4, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGoals - May 2015\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\nGoals\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nMay 1, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBLASTN - C.gigas OA Larvae to C.gigas Ensembl 1.24 BLAST DB\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 29, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation – Geoduck Gonad in Paraffin Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer Data - Geoduck RNA from Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nApr 24, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation – Geoduck Gonad in Paraffin Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nApr 23, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer Submission - Geoduck Gonad RNA from Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nApr 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nQuality Trimming - C.gigas Larvae OA BS-Seq Data\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nQuality Trimming - LSU C.virginica Oil Spill MBD BS-Seq Data\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequence Data Analysis - LSU C.virginica Oil Spill MBD BS-Seq Data\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequence Data Analysis - C.gigas Larvae OA BS-Seq Data\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequence Data - C.gigas OA Larvae BS-Seq Demultiplexed\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequence Data - LSU C.virginica Oil Spill MBD BS-Seq Demultiplexed\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Geoduck Gonad in Paraffin Histology Blocks\n\n\n\n\n\n\n\n2015\n\n\nProtein expression profiles during sexual maturation in Geoduck\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing Data - C.gigas Larvae OA\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 19, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEpinext Adaptor 1 Counts - LSU C.virginica Oil Spill Samples\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 17, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTruSeq Adaptor Counts – LSU C.virginica Oil Spill Sequences\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 16, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTruSeq Adaptor Identification Method Comparison - LSU C.virginica Oil Spill Sequences\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Claire’s C.gigas Sheared DNA\n\n\n\n\n\n\n\n2015\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nMar 3, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Quality Assessment - C.gigas OA larvae Illumina libraries\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBS-seq Library Prep - C.gigas Larvae OA 1000ppm\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 27, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - C.gigas Larvae 1000ppm\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 26, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Claire’s Sheared C.gigas Mantle Heat Shock Samples\n\n\n\n\n\n\n\n2015\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nFeb 26, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - C.gigas Sheared DNA from 20140108\n\n\n\n\n\n\n\n2015\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nFeb 20, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Prep - Quantification of C.gigas larvae OA 1000ppm library\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing Data - LSU C.virginica MBD BS-Seq\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisulfite NGS Library Prep - Bisulfite Conversion & Illumina Library Construction of C.gigas larvae DNA\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nFeb 6, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisuflite NGS Library Prep – C.gigas larvae OA bisulfite library quantification\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 28, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisuflite NGS Library Prep - C.gigas larvae OA bisulfite DNA (continued from yesterday)\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 27, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisuflite NGS Library Prep - C.gigas larvae OA bisulfite DNA\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 26, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Cleanup - LSU C.virginica MBD BS Library\n\n\n\n\n\n\n\n2015\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Bisulfite Conversion - C.gigas larvae OA Sheared DNA\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSpeedVac - C.gigas larvae OA DNA\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas larvae from 2011 NOAA OA Experiment\n\n\n\n\n\n\n\n2015\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2015\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisulfite NGS Library - LSU C.virginica Oil Spill MBD Bisulfite DNA Sequencing Submission\n\n\n\n\n\n\n\n2014\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisulfite NGS Library Prep - LSU C.virginica Oil Spill MBD Bisulfite DNA and Emma’s C.gigas Larvae OA Bisulfite DNA (continued from yesterday)\n\n\n\n\n\n\n\n2014\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisulfite NGS Library Prep - LSU C.virginica Oil Spill Bisulfite DNA and Emma’s C.gigas Larvae OA Bisulfite DNA\n\n\n\n\n\n\n\n2014\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBisulfite Conversion - LSU C.virginica Oil Spill MBD DNA and Emma’s C.gigas Larvae OA DNA\n\n\n\n\n\n\n\n2014\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 16, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Claire’s C.gigas Female Gonad for Illumina Bisulfite Sequencing\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEtOH Precipitation - LSU C.virginica Oil Spill MBD Continued (from 20141126)\n\n\n\n\n\n\n\n2014\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nDec 2, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMethylated DNA Enrichment (MBD) - LSU C.virginica Oil Spill gDNA\n\n\n\n\n\n\n\n2014\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 26, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing - LSU C.virginica Oil Spill gDNA\n\n\n\n\n\n\n\n2014\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGel - Sheared gDNA\n\n\n\n\n\n\n\n2014\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Seq - C.gigas Total RNA from Claire’s Pre/Post Heat Shock\n\n\n\n\n\n\n\n2014\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas Larvae from Emma OA Experiments\n\n\n\n\n\n\n\n2014\n\n\nCrassostrea gigas larvae OA (2011) bisulfite sequencing\n\n\n\n\n\n\n\n\n\n\n\nNov 21, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRAD Sequencing - Oly Oyster gDNA-01 RAD Library (from 20141110)\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nNov 17, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Oly Oyster gDNA-01 RAD Library\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 10, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLibrary Prep - Oly Oyster gDNA-01 RAD\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 7, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Shearing & Size Selection - Oly Oyster gDNA RAD P1 Adapters (from 20141105)\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 6, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLigation - Illumina P1 Adapters for Oly Oyster gDNA-01 RAD Sequencing (from 20141031)\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nNov 5, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digest - Oly Oyster gDNA-01 for RAD Sequencing (from 20141029)\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 31, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Allocation - Oly Oyster gDNA-01 for RAD Sequencing (from 20141022)\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 29, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Oly Oyster gDNA-01 for RAD Sequencing (from 20141014)\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 22, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Olympia Oyster Populations for RAD Sequencing\n\n\n\n\n\n\n\n2014\n\n\nOlympia oyster reciprocal transplant\n\n\n\n\n\n\n\n\n\n\n\nOct 14, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPackage - Received Package from Jerome LaPeyre from LSU\n\n\n\n\n\n\n\n2014\n\n\nLSU C.virginica Oil Spill MBD BS Sequencing\n\n\n\n\n\n\n\n\n\n\n\nSep 26, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas Larvae from Katie Latterhos\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 23, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas Larvae from Katie Latterhos and Emma\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 22, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Mac’s Bisulfite-Treated DNA\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 3, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Mac’s Bisulfite-Treated DNA\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 28, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA-Seq - Sea Star Data Download\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nMay 28, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Jessica’s Geoduck Larval Stages\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Mackenzie’s C.gigas EE2 Gonad Samples\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 16, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - Colleen Sea Star (Pycnopodia) Coelomycete RNA for Illumina Sequencing\n\n\n\n\n\n\n\n2014\n\n\nSamples Submitted\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Colleen Sea Star (Pycnopodia) Coelomycete Sample\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Colleen Sea Star (Pycnopodia) Coelomycete Samples\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 28, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Claire’s C.gigas Female Gonad\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nApr 25, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation - Colleen’s Sea Star Coelomycete RNA from Yesterday\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 25, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Clean Up - Colleen’s Sea Star Coelomycete RNA from 20140416\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 24, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Colleens’ Sea Star Coelomycetes Samples\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 16, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Test Sample\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 16, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPhenol-Chloroform DNA Clean Up - Mac and Claire’s Samples (from 20140410)\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Sea Star Coelomocytes (from Colleen)\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Claire’s C.gigas Female Gonad and Mac’s C.gigas Gonad\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nCloned Hard Drive - Windows XP Opticon Computer (Aquacul8)\n\n\n\n\n\n\n\n2014\n\n\nComputer Servicing\n\n\n\n\n\n\n\n\n\n\n\nApr 4, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA gel - Claire’s C.gigas Female Gonad and Mac’s C.gigas Gonad\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nApr 4, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Sea Star Coelomocytes (provided by Colleen Burge)\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 2, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Sea Star Coelomocytes (provided by Colleen Burge)\n\n\n\n\n\n\n\n2014\n\n\nSea star RNA-seq\n\n\n\n\n\n\n\n\n\n\n\nApr 1, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Mackenzie’s C.gigas Gonad Sample\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 1, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - C.gigas Female Gonads (from frozen)\n\n\n\n\n\n\n\n2014\n\n\nLineage-specific DNA methylation patterns in developing oysters\n\n\n\n\n\n\n\n\n\n\n\nMar 28, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quality Check - Yanouk’s Oyster gDNA\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 19, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Precipitation - Geoduck DNA from 20140213\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 18, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 13, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Geoduck\n\n\n\n\n\n\n\n2014\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 12, 2014\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGylcogen Assay - Emma’s C.gigas Whole Body Samples (continued from yesterday)\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 7, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGylcogen Assay - Emma’s C.gigas Whole Body Samples\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 6, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGylcogen & Carboyhydrate Assays - Emma’s C.gigas Whole Body Samples (continued from yesterday)\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 26, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGylcogen and Carboyhydrate Assays - Emma’s C.gigas Whole Body Samples\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Lake Trout C1q\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 28, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Lake Trout C1q\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 10, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Quantification - Yanouk’s DNA Samples\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 4, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Lake Trout C1q\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 19, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Hexokinase Partial CDS\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Hexokinase and Partial Exon #1\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Hexokinase Promoter and CDS (repeat from 20130227)\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Hexokinase Promoter and CDS\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 27, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Herring RNA from 20091026\n\n\n\n\n\n\n\n2013\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 13, 2013\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Halley cDNA Check\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 28, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - FISH441 RNA\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 21, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Manila Clam Larvae cDNA (from August 2012 - Dave’s Notebook)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 8, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Manila Clam Larvae cDNA (from August 2012 - Dave’s Notebook)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 8, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Manila Clam Larvae cDNA (from August 2012 - Dave’s Notebook)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 8, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased Manila Clam Larvae RNA (from August 2012 - Dave’s Notebook)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 26, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReceived oysters from Taylor Shellfish.\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 16, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Opticon Test\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 10, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMinipreps - Emma’s Illumina Library Cloning\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nIllumina RNAseq Library Construction - 32 C.gigas Individuals\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nOligo Reconstitution - Illumina RNAseq Library Oligos and Barcodes\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nChloroform Clean Up - Lexie’s QPX RNA from 20110504\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - QPX RNA and DNA for Illumina 36bp single-end RNA/DNAseq\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nQPX Sample Pooling for Illumina Sequencing\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Detection of V.tubiashii Presence and Expression Using VtpA Primers in DNA/cDNA from yesterday\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 1, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - DNased C.gigas Larval RNA from 20120427\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\nM-MLV\n\n\nreverse transcription\n\n\nRNA\n\n\nVibrio tubiashii\n\n\n\n\n\n\n\n\n\n\n\nApr 30, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Check DNased RNA from Earlier Today for Residual gDNA\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 27, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - C.gigas Larvae RNA from yesterday\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 27, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - C.gigas Larvae from Taylor Summer 2011\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - C.gigas Larvae from Taylor Summer 2011\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Taylor Water Filter DNA Extracts from 20120322 - Sam White\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 26, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Repeat of qPCR from Earlier Today\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 23, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Taylor Water Filter DNA Extracts from Yesterday\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 23, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Extraction - Taylor Water Filter Samples from 2011\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 22, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Dave’s Manila Clam (Venerupis philippinarum) DNased RNA from 20120307 and 20120302\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 12, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Dave’s Manila Calm (Venerupis philippinarum) DNased RNA from yesterday and 20120302\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 8, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Dave’s Manila Clam (Venerupis philippinarum) Gill Samples (#25-48)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 7, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - Dave’s Manila Clam (Venerupis philippinarum) Gill RNA from Yesterday\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Dave’s Manila Clam (Venerupis philippinarum) Gill Samples (#1-24)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - cDNA from 20120208\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - cDNA from earlier today\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 8, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - C.gigas larvae DNased RNA (from 20120125)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 8, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased RNA from earlier today\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 25, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNAse - C.gigas RNA from 20120124\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 25, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - C.gigas Larvae from 20110412 & 20110705 (Continued from 20120112)\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 24, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - C.gigas Larvae from 20110412 & 20110705\n\n\n\n\n\n\n\n2012\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2012\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - COX/PGS Clones from yesterday/today\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 14, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMini-preps - COX/PGS Cloning Colonies from today\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 13, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - COX/PGS Cloning Colony Screens from yesterday\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 13, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nCloning - Purified COX/PGS “qPCR Fragment” from 20111006\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 12, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Purified COX/PGS 1/2 DNA from earlier today\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 6, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Region Outside of COX/PGS qPCR Primers\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 6, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation - Full-length PGS1 cDNA (from 20110921)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 29, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Full-length PGS1 cDNA\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 21, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation - Purified PGS1 PCR from yesterday\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 21, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Full-length PGS1 cDNA\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 20, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Full-length PGS2 cDNA\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 14, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Full-length PGS1 & PGS2 cDNAs\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas V.vulnificus Exposure cDNA (from 20110311)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - C.gigas COX2/PGS2 Clone #4 from 20110728\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 4, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPlasmid Isolation & Sequencing - C.gigas COX2/PGS2 Clones (from yesterday)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 28, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacterial Cultures - C.gigas COX2/PGS2 Clones (from yesterday)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 27, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nColony PCRs - C.gigas COX2/PGS2 Clones (from yesterday)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 26, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nCloning - C.gigas COX2/PGS2 5’/3’ RACE Products (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\n5’/3’ RACE - C.gigas COX2/PGS2 Nested RACE PCR\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\n5’/3’ RACE - C.gigas COX2/PGS2 RACE PCR\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 22, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - PGS Hi 4 (PGS2/COX2)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 15, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPlasmid Isolation - Miniprep on PGS Hi 4 Colony from yesterday\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 12, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Colony PCR on Restreaked PGS2 Clones from 20110707\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacterial Cultures - Liquid Cultures of PGS2/COX2 Colonies from 20110707\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nClone Restreaking - PGS2 Hi/Lo Clones (from 20110421)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 7, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas GAPDH second rep on V.vulnificus exposure cDNA (from 20110311) and standard curves for COX1, COX2, GAPDH\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 3, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas actin and GAPDH on V.vulnificus exposure cDNA (from 20110311)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 2, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas 18s and EF1a on V.vulnificus exposure cDNA (from 20110311)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 2, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas COX2 on V.vulnificus exposure cDNA (from 20110311)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas COX1 on V.vulnificus exposure cDNA (from 20110311)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Hard Clam NGS Primer Checks\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 23, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Lexie’s QPX Temp & Tissue Experiment (see Lexies Notebook 4/26/2011)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 20, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Emma’s New 3KDSqPCR Primers\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 20, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Hard Clam Gill DNased RNA (from 20110509)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - Hard Clam Gill RNA (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 9, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Hard Clam Gill Tissue from Vibrio Experiment (see Dave’s Notebook 5/2/2011)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 9, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPrimer Design - Hard Clam NGS Primers\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Hard Clam Gill Tissue from Vibrio Experiment (see Dave’s Notebook 5/2/2011)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 6, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReceived - Live Hard Clams From Scott Lindell\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 28, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacterial Cultures - Colonies Selected from Yesterday’s Colony PCRs\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMini Preps - Liquid Cultures from yesterday\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 26, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacterial Cultures - Colonies Selected by Steven from Steven’s Re-Streaked Plate\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nColony PCR - Colonies from COX1 Genomic Cloning (from 20110411)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nColony PCR - 5’RACE Colony: COX2 (repeat of yesterday’s PCR)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 22, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nColony PCR - 5’ RACE Colony: COX2\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 21, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nColony PCR - 5’ RACE Colonies\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nLigations - COX1/COX2 PCR Products\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\n5’/3’ RACE PCRs - Nested PCRs for COX2 Sequence\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 7, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\n5’/3’ RACE PCRs - COX2 Sequence on 5’ & 3’ RACE Libraries (from 20080619)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 6, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas BB/DH cDNA for PROPS (TIMP3(BB) primers)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMar 21, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas BB/DH cDNA for PROPS (HMGP primers)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMar 21, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas COX1/COX2 Tissue Distribution\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 15, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - C.gigas BB/DH cDNA for PROPS\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMar 14, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - C.gigas BB/DH DNased RNA (from 20090507) for PROPS\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMar 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - C.gigas DNased RNA (from 20110131) from V.vulnificus Exposure & Tissues (from 20110111)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD Sequencing Submission\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 10, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Pooled Black Abalone Dg RNA (from Abalone Dg Exp 1)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 7, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\n3’RACE - C.gigas 3’RACE for COX2\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 4, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoDrop1000 Comparison - Roberts vs. Young Lab\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Check DNased RNA BB01 for Residual gDNA (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - C.gigas BB01 (PROPS) RNA (from 20090507)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMar 1, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Check DNased RNA BB01 for Residual gDNA (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - C.gigas BB01 from 20110225\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 28, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precpitation - DNased RNA BB01 (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Check DNased RNA BB01 for Residual gDNA (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - C.gigas BB01 from 20110216\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 25, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEthanol Precipitation - DNased RNA BB01 (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 16, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Check DNased RNA BB01 & 09 (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 16, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - C.gigas BB/DH (PROPS) RNA (from 20090507)\n\n\n\n\n\n\n\n2011\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nFeb 16, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nData Analysis - Young Lab ABI 7300 Calibration Checks\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Test Young Lab qPCR Calibration\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoDrop1000 Comparison - Roberts vs. Young Lab\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 9, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Test Young Lab qPCR Calibration (Repeat)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 8, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - New C. gigas COX Primers for Sequencing of Isoforms\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 8, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Test Young Lab qPCR Calibration (Repeat)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 8, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Test Young Lab qPCR Calibration (Repeat)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 4, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Test Young Lab qPCR Calibration\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 4, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic PCR - Repeat of C.gigas COX genomic PCR from 20110118\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 31, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - DNase C.gigas RNA from 20110120, 20110121 and 20110124\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 28, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Various C.gigas Tissue from 20110111\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 24, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Various C.gigas Tissue from 20110111\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 21, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Various C.gigas Tissue from 20110111\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 20, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGenomic PCR - C.gigas cyclooxygenase (COX) genomic sequence\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 18, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacterial Dilutions - Determination of Colony Forming Units from Gigas Bacterial challenge (from earlier today)\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGigas Bacterial Challenge - 1hr & 3hr Challenges with Vibrio vulnificus\n\n\n\n\n\n\n\n2011\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2011\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD - Retrieved SOLiD Library Samples from CEG from 20101213\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 23, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - COX qPCR Vibrio Exposure Response Check\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 13, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - COX qPCR Primer Test and Tissue Distribution\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 10, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digestions/Ligations - MS-AFLP\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 30, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digestions - HpaII and MspI on Mac’s C.gigas Samples: Round 1\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 29, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEtOH Precipitations - HpaII and MspI 2nd Round Digests from 20101124\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 29, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digestions - HpaII and MspI on Mac’s C.gigas Samples: Round 2\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPhenol:Chloroform Extractions and EtOH Precipitations - HapII and MspI digests from yesterday\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 23, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digestions - HpaII and MspI on Mac’s C.gigas gDNA Samples: Round 1\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 22, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nQPX Washes\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 26, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 26, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEtOH Precipitation - Whale gDNA from 20101022\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 25, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 22, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReceived Hard Clam Samples and Live Clams from MBL\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 19, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReceived Hard Clam Samples from Rutgers\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 15, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Test Plate for Opticon 2\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 12, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nOpticon Calibration\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 17, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Hard Clam Primers on cDNA from yesterday\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\nBigDef\n\n\nC1qTNG\n\n\nCA\n\n\nCFX96\n\n\nCytP450-like\n\n\nferritnin\n\n\ngraphs\n\n\nGST\n\n\nHard clam\n\n\nHemoDef\n\n\nHSP 70\n\n\nImmomix\n\n\nlysozyme\n\n\nMA\n\n\nMAX\n\n\nMercenaria mercenaria\n\n\nMercenaria_Rel\n\n\nmetallothionein\n\n\nMm_TRAF6\n\n\nNADHIV\n\n\nqPCR\n\n\nRACK1\n\n\nSenescentProt\n\n\nSTI\n\n\nSYTO 13\n\n\nTfAP1\n\n\nTLR\n\n\n\n\n\n\n\n\n\n\n\nSep 9, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - DNased Hard Clam RNA from earlier today\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase - DNasing Hard Clam RNA from yesterday\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEtOH Precipitation - Hard Clam RNA from earlier today\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 7, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Hard Clam Tissues Rec’d from Rutgers on 20100820\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 7, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPackage - Hard Clam Samples from MBL\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 2, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPackage - Hard Clam Samples from Rutgers\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 20, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - HpaII/MspI Digests from earlier today\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digests - Various gigas gDNA from earlier today\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Various gigas samples (continued from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Various gigas samples\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMeDIP - SB/WB Fragmented gDNA EtOH precipitation (continued from 20100702)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMeDIP - SB/WB Fragmented gDNA (continued from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRestriction Digests - Various gigas gDNAs of Mac’s\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMeDIP - SB/WB Fragmented gDNA (continued from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 1, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMeDIP - SB/WB Fragmented gDNA (from 20100625)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - Fragmented SB/WB gDNA (from 20100625)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Sonication - SB/WB gDNA pools (prep for MeDIP) from 20100618\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSamples Received - Hard Clam samples from Rutgers and MBL\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 24, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Sonication - SB/WB gDNA pools (prep for MeDIP) from earlier today\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 18, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Precipitation - SB/WB gDNA pools (prep for MeDIP)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 18, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Mac gigas larvae samples: control larvae 6.7.10 and 5-aza tr larvae 6.7.10\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Mac gigas gill samples (continued from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Mac gigas gill samples\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\nCrassostrea gigas\n\n\nDNA Isolation\n\n\nDNazol\n\n\ngill\n\n\nPacific oyster\n\n\nR37\n\n\nR51\n\n\nRNAlater\n\n\n\n\n\n\n\n\n\n\n\nJun 4, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Mac gigas gill samples (continued from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 28, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPackage Rec’d - From NOAA in Connecticut\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\nalgae\n\n\nIsochrysis sp. T-150\n\n\nNOAA\n\n\npackage\n\n\nTetraselmis cheri Ply429\n\n\nThalassiaosira weissflugii TW\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Mac gigas gill samples\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\nCrassostrea gigas\n\n\nDNazol\n\n\nPacific oyster\n\n\nR51\n\n\nRNAlater\n\n\n\n\n\n\n\n\n\n\n\nMay 27, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD ePCR/Templated Bead Prep - Lake Trout Lean library\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 26, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD Templated Bead Prep - Yellow perch CT, WB and lake trout Lean libraries (continued from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 13, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD ePCRs - Yellow perch CT, WB and lake trout Lean libraries\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 12, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPackage - Hard Clam gill tissue/hemolymph in RNA later\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 5, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Qiagen Kit Comparison\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\nDNeasy\n\n\ngDNA\n\n\ngel\n\n\nHyperladder I\n\n\ntroubleshooting\n\n\n\n\n\n\n\n\n\n\n\nMay 5, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - V.tubiashii primers test (Vpt A and Vt IGS)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 3, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTemplated Bead Prep SOLiD Libraries - Yellow perch WB, lake trout Lean and Sisco, and herring G/O HWS09 libraries\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 16, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nePCR SOLiD Libraries - Lake Trout Sisco and Herring G/O HPWS09 libraries (from 20100408)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTemplated Bead Prep SOLiD Libraries - Abalone CC, CE pools and yellow perch CT, PQ libraries\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nePCR SOLiD Libraries - Yellow perch PQ, WB and Lake Trout Lean libraries (from 20100408)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nePCR SOLiD Libraries - Abalone CC, CE pools and yellow perch CT SOLiD libraries (from 20100408)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 12, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ncDNA clean up & Bioanalyzer for SOLiD Libraries - Abalone, Yellow Perch, Lake Trout, Herring\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nGel Purification & PCR cDNA SOLiD Libraries - Abalone, Yellow Perch, Lake Trout, Herring\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 7, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription SOLiD Libraries - Abalone, Yellow Perch, Lake Trout, Herring\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 6, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHibridizaton/Ligation SOLiD Libraries - Abalone, Yellow Perch, Lake Trout, Herring\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 5, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer Total, mRNA and post-fragmentation SOLiD Libraries - Abalone pools\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 2, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation and Fragmentation for SOLiD Libraries - Pooled abalone mRNA (from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 1, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation & mRNA Isolation for SOLiD Libraries - Pooled abalone total RNA: Carmel control, Carmel exposed\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 31, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer for SOLiD Libraries - Fragmented mRNA from Perch, Lake Trout & Herring RNA samples\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 29, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD Library Prep - mRNA (perch, lake trout, herring from 20100318) Fragmentation\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 25, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer for SOLiD libraries - Total and mRNA from Perch, Lake Trout & Herring RNA samples (CONTINUED from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 18, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Precipitation for SOLiD - Perch, Lake Trout, & Herring mRNA (CONTINUED from yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 17, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation for SOLiD - Perch, Lake Trout, and Herring total RNA\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 16, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Test Lexie’s Mercenaria 18s contamination issue\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 10, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Mac’s BB/DH cDNA from 20091223\n\n\n\n\n\n\n\n2010\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nJan 15, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD Bead Titration - Herring fragmented cDNA libraries: 2LHKOD09, 4LHTOG09, 6LHPWS09\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD ePCRs - Herring cDNA libraries\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 13, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD ePCR - Herring fragmented cDNA library: 2LHKOD09\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - Mac’s BB/DH cDNA from 20091223\n\n\n\n\n\n\n\n2010\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nJan 11, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD Bead Titration - Herring fragmented cDNA library 3LHSITK09 (CONTINUED from ePCR yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD ePCR - Herring fragmented cDNA library: 3LHSITK09\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 7, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD Bead Titration - Herring fragmented cDNA library 3LHSITK09(CONTINUED from ePCR yesterday)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 7, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - Tim’s Adult Gigas gill cDNA (from 20091009)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 6, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSOLiD ePCR - Herring fragmented cDNA library: 3LHSITK09 (from 20091209)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 6, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - Tim’s Adult Gigas gill cDNA (from 20091009)\n\n\n\n\n\n\n\n2010\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 5, 2010\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - BB & DH cDNA (from 20091223)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 31, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - BB & DH cDNA (from 20091223)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 30, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - BB & DH cDNA (from 20091223)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAlaska sockeye salmon sampling (with Seebs): Family #13\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - BB & DH cDNA (from 20091223) and Emma primer sets for testing\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - BB & DH cDNA (from 20091223)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 28, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - BB & DH cDNA (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - Mac methylation samples, Sam rhodopsin samples, Lisa samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - BB & DH cDNA (from earlier today)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - BB & DH DNased RNA (from 20090514)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nDec 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Sepia cDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Sepia cDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Sepia cDNA and DNased RNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Sepia cDNA (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Abalone 07:12 DNased RNA (from 20090623)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Sepia DNased RNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer - Herring Liver cDNA for SOLiD Libraries\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 11, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEmulsion PCR - Herring Liver cDNA for SOLiD Libraries\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Herring Liver mRNA for SOLiD Libraries\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Adapter Hybridization and Ligation - Herring Liver mRNA for SOLiD Libraries\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Fragmentation - Herring Liver mRNA for SOLiD Libraries\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Sepia samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 4, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - Dungan Isolates, Lake Trout HRM and Emma DD cloning\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 3, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Precipitation - Herring Liver mRNA for SOLiD Libraries (continued from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 3, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHard Clam Challenge - QPX Strain S-1 (continued from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation - Herring Liver RNA for SOLiD Libraries (continued from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation - Herring Liver RNA for SOLiD Libraries\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHard Clam Challenge - QPX Strain S-1\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBL Shipment - Hard Clam gill tissue in RNA Later\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBL Shipment - Sepia tissue samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 25, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBL Shipment - Hard Clams\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHerring 454 Data\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 19, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMBL Shipment - MV oysters/cod\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 19, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - “Unknown” Dungans/Lyons\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 18, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - “Unkown” Dungans/Lyons\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 17, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHard Clams - Shipment from Rutgers\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - Lake Trout HRM\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nOyster CO2/Mechanical Stress - Water quality\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submissions to MoGene for 454 Analysis - Herring Liver and Testes mRNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\nSamples Submitted\n\n\n\n\n\n\n\n\n\n\n\nOct 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Herring gonad/ovary RNA (from 20091023)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation - Herring gonad/ovary RNA (from 20091023)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 26, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Herring Liver RNA (from 20091021)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 26, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation - Herring Liver Samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Herring Gonad/Ovary Samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Herring Gonad/Ovary Samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Herring Liver Samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 21, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Herring Liver Samples (LHPWS09 1-6)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 20, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - Tim’s adult gigas challenge cDNA (from 20091009)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Tim’s adult gigas challenge cDNA (from 20091009)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Tim’s adult gigas challenge cDNA (from today)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Tim’s adult gigas challenge DNased RNA (from 20091008)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Tim’s adults gigas challenge re-DNased RNA (from today)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Re-DNase of Tim’s adult gigas challenge RNA (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Tim’s adults gigas challenge DNased RNA (from today)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Tim’s adults gigas challenge DNased RNA (from today)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Tim’s adult gigas challenge RNA (from 20090930)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer Submission - Trout RBC, Colleen’s gigas GE sample, Mac’s DH/BB PCR for SOLiD WTK\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription/cDNA purification/Emulsion PCR - Ligation rxns of trout fragmented RNA for SOLiD WTK (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nAdapter Ligation - Rick’s trout fragmented control/poly I:C samples for SOLiD WTK\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Tim’s adults gigas challenge DNased RNA (from 20091002)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Tim’s adult gigas challenge RNA (from 20090930)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - Check gDNA contamination with EF1 & 18s primers in gigas gill RNA (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nOct 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Tim’s adult gigas challenge samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 30, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBioanalyzer Submission - Rick’s trout RBC samples (various dates)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Fragmentation - Rick’s trout RBC samples prepped earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEtOH Precipitation - Rick’s trout Ribosomoal-depleted RNA for SOLiD WTK (from today)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRibosomal-depleted RNA - Rick’s trout RBC samples for the SOLiD WTK\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 28, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Fragmentation - Rick’s trout RBC samples prepped earlier today (see below)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Rick’s trout RBC samples previously treated with Ribominus Kit (by Mac)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Gigas BB and DH samples previously treated with Ribominus Kit (by Mac)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 18, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Gigas BB and DH samples previously treated with Ribominus Kit (by Mac)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHRMs - Lake Trout SNPs (HRM_white-05 & HRM_white_06)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHRMs - Lake Trout SNPs (HRM_white-03 & HRM_white-04)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHRM - Lake Trout SNPs (HRM_white-02)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nHRM - Lake Trout SNPs (HRM-white-01)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 3, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - HRM Lake Trout SNP primer test\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nSep 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPrimers - Lake Trout Primers for HRM\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Gigas gDNA test of recalibrated Opticon 2\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Recalibration of Opticon 2\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 26, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Precipitation - C.pugetti DNA for JGI submission (continued from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 26, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Precipitation - C.pugetti DNA for JGI submission\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 25, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Additional Calibration test of Opticon 2\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Carita Primer Test for High Resolution Melt (HRM) Curve Analysis\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 17, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Calibration test of Opticon 2\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Calibration test of Opticon 2\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nAug 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C.pugetti culture (from 20090713)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Gigas DNA for Opticon testing\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRT Rxns - H.crach DNased RNA (from 20090623)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nNanoDrop - H.crach DNased RNA (from 20090623)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 21, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone cDNA (07:12 set from 3/3/2009 by Lisa) and DNased RNA (from 20090623)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 20, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone gDNA (H.crach 06:7-1)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 17, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone cDNA (07:12 set from 3/3/2009 by Lisa) and DNased RNA (from 20090623)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone cDNA (07:12 set from 3/3/2009 by Lisa) and DNased RNA (from 20090623)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone gDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone RNA/DNased RNA & “dirty” and “clean” cDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Dungan isolate (MIE-14v) gDNA from 20090708\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C.pugetti large culture\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone gDNA/cDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 11, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased Abalone Dg RNA from 20090625\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Precipitation CONTINUED - Dungan MIE-14v gDNA from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Bay/Sea Scallop DNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased Abalone Dg RNA from 20090625\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Precipitation - Dungan MIE-14v gDNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Dungan isolate MIE-14v\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSpec Reading - C.pugetti gDNA from 20090526\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti liquid cultures\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJul 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 30, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 26, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone Dg DNased RNA from yesterday and earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg RNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 25, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCRs - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg RNA (07:12 set)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEtOH Precipitation - DNased Abalone Dg RNA from yesterday AND the 07:12 set (DNased by Lisa 20090306)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased Abalone Dg RNA from earlier today AND the 07:12 set (DNased by Lisa 20090306)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg DNased RNA 20090610\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 18, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from 20090614\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 17, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA: Test Immomix (SYTO13) vs. Strategene SYBR\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - MV hemocyte cDNA from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - MV hemocyte DNased RNA from 20090612\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased MV hemocyte RNA from earlier today AND Turbo kit test\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - MV hemocyte RNA from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - MV hemocyte RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 11, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Martha’s Vineyard (MV) hemocytes\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 11, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Re-DNased abalone Dg RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Re-DNased abalone Dg RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg DNased RNA from 20090605\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - C.pugetti gDNA from 20090526\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - C.pugetti gDNA from 20090513 & 20090526\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Re-DNased abalone Dg RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg DNased RNA from 20090605\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C.pugetti culture (1x 1L)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased abalone Dg RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg RNA isolated yesterday and from 20090518\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Abalone Dg Project Samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 4, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Abalone Dg RNA isolated yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 3, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - C.pugetti DNA from 20090513 & 20090526\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Abalone Dg Project\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - C.pugetti DNA from 20090513 & 20090526\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Gel - JGI QC check of C. pugetti DNA from 20090526\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJun 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - C. pugetti (from 20090518)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 26, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Methylation Test - Gigas site gDNA (BB & DH) from 20090515\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 19, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Abalone Dg Project samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 18, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nC.pugetti - Liquid Cultures\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 18, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Gigas Dermo Samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Mac’s BB and DH site samples\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - Mac’s gigas DNased RNA from 20090512\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - C.pugetti\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Mac’s gigas DNased RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment (Rigorous!) - Mac’s gigas RNA/Re-DNased RNA from 20090507 & 20090508, respectively\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNA Isolation - Mac’s gigas samples from 20090505 & 20090506\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 11, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Re-DNased oyster RNA from today\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Oyster RNA from yesterday\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - DNased oyster RNA from earlier today\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nDNase Treatment - Oyster RNA from today\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Mac’s oyster tissues (BB and DH) (CONTINUED from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Mac’s oyster tissues (BB and DH) (CONTINUED from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti liquid cultures\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Mac’s oyster tissues (BB and DH)\n\n\n\n\n\n\n\n2009\n\n\nPROPS\n\n\n\n\n\n\n\n\n\n\n\nMay 5, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti liquid cultures\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMay 1, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti plate (from 20090424)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 30, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti plate (from 20090424)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 29, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti liquid culture\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugetti plate\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - cDNA from DNased Abalone RNA from 20090420\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone DNased RNA from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 21, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugettii culture CONTINUED (from 20090419)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 21, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Removal - Abalone RNA from 20090402 and 20090331\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 20, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBacteria - C. pugettii culture\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 19, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - Dungan isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 17, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Two new Dungan isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Rab7_SYBR primers on abalone RNA and DNased RNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Two new Dungan isolates from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Two new Dungan isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 15, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Test QT Kit with No RT Abalone rxns from 20090408\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Bay/Sea scallop gDNAs\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Bay/Sea scallop gDNA isolated earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - Bay/Sea Scallop and hybrid samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Abalone gDNA/RNA/cDNA w/new TOLLIP primer\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Old Dungan isolates #1-35 w/EukA/B primers\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Bay/Sea scallop hybrids\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 10, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Dungan isolates from 20090402 with Euk primers\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Check DNased abalone RNA (by Lisa) for gDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Repeat (modified) of yesterday’s abalone cDNA check\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone cDNA (QT) from earlier today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ncDNA - Abalone RNA from 20090331 & 20090402\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - New Dungan isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - New Dungan isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 3, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Abalone RNA, check for gDNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 3, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\ngDNA Isolation - New Dungan isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Abalone digestive gland samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nApr 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Abalone digestive gland samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 31, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Abalone digestive gland samples\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Virginica cDNA (see workup sheet below for more info regarding cDNA)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Sample Submission Hard clam gill #1 mRNA from 20090313\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - hard clam gill #1 continued from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - hard clam gill #1 DNased RNA from today\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Precipitation - Hard clam gill #1 RNA from 20080819\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Repeat of qPCR from earlier today with fresh primer working stocks\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Repeat of 20090227 qPCR with clean water\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nMar 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - New 16s primers for V.tubiashii Control vs. Autoclaved gigas samples (see 20090224)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 27, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - Replicate of V.tubiashii Control vs. Autoclaved gigas samples (see yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 25, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nqPCR - V.tubiashii Control vs. Autoclaved gigas samples (see below)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nReverse Transcription - V.tubiashii DNAsed RNA (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 24, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - V.tubiashii samples from autoclaved gigas exposure (from 20081218)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nEpigenetics Experiment - Gigas treatment\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 20, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSample Submission - V.tubiashii Mass Spec\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 18, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrypsin digestion - Vibrio 2D spots CONTINUED (from yesterday)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nTrypsin digestion - Vibrio 2D spots from 20081217\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 11, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA - Submission for Agilent Bioanalyzer\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nFeb 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA - Precipitation continued from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 23, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nmRNA Isolation - Hard Clam gill and hemo RNA\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA - Hard clam hemo RNA (from 20090121)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 22, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Hard clam hemo (from 20090121)\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 21, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBleeding/Tissue Collection - Hard Clams\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 20, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA - Precipitation of Hard Clam Hemo RNA from 20090116\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 20, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Hard clam gill, hemos\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 16, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBleeding - Hard Clams\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA - Precipitation continued from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 14, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA - Reprecipitation of hard clam RNA from yesterday\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 13, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBleeding - Hard Clams\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - Hard Clam hemolymph from 20090108, 20090109\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 12, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBleeding - Hard Clams\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 9, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nBleeding - Hard Clams\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 8, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Dungan Isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 7, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nPCR - Dungan Isolates\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 6, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE, Western Blot - Test of HSP70 Ab on heat stressed shellfish for FISH441\n\n\n\n\n\n\n\n2009\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nJan 2, 2009\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE, Western Blot - Test of new Western Breeze Kit & HSP70 Ab for FISH441\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\nanti-HSP70\n\n\nanti-myc\n\n\nantibody\n\n\nCoomassie\n\n\nCrassostrea gigas\n\n\nMSTN\n\n\nMSTN1b\n\n\nmyostatin\n\n\nPacific oyster\n\n\nprotein\n\n\nSDS-PAGE\n\n\nSeeBlue Plus\n\n\nWestern blot\n\n\nWestern Breeze Chromogenic (anti-mouse) Kit\n\n\n\n\n\n\n\n\n\n\n\nDec 31, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Gel - V. tubiashii mRNA samples (from 20081224)\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 29, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nrRNA Removal - V. tubiashii total RNA from yesterday\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 24, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nRNA Isolation - V. tubiashii from challenge (see 20081216)\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\nMICROBExpress Kit\n\n\nNanoDrop1000\n\n\nRNA isolation\n\n\nRNA quantification\n\n\nTriReagent\n\n\nVibrio tubiashii\n\n\n\n\n\n\n\n\n\n\n\nDec 22, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nVibrio challenge CONTINUED (from yesterday)\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 19, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nVibrio challenge CONTINUED (from yesterday)\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 18, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nVibrio challenge CONTINUED (from yesterday)\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 17, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nVibrio challenge\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\nbacterial challenge\n\n\nbacterial culture\n\n\nVibrio exposure\n\n\nVibrio tubiashii\n\n\n\n\n\n\n\n\n\n\n\nDec 16, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE/Western - anti-HSP70 Ab Re-test\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\nanti-HSP70\n\n\nantibody\n\n\nchemiluminescent\n\n\nCoomassie\n\n\nCrassostrea gigas\n\n\nhemocyte\n\n\nhemolymph\n\n\nPacific oyster\n\n\nprotein\n\n\nSDS-PAGE\n\n\nSeeBlue Plus\n\n\n\n\n\n\n\n\n\n\n\nDec 16, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE/Western - Attempt to fix/identify problem(s) with Westerns\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 12, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE/Western - anti-HSP70 Ab test CONTINUED (from yesterday)\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nDec 10, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE/Western - anti-HSP70 Ab test\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\nanti-HSP70\n\n\nantibody\n\n\nCoomassie\n\n\nCrassostrea gigas\n\n\ngill\n\n\nmucus\n\n\nOctopus rubescans\n\n\nPacific oyster\n\n\nprotein\n\n\nred octopus\n\n\nSDS-PAGE\n\n\nskin\n\n\n\n\n\n\n\n\n\n\n\nDec 9, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSDS/PAGE/Western - Purified (His column) FST samples from 20081112\n\n\n\n\n\n\n\n2008\n\n\nMiscellaneous\n\n\n\n\n\n\n\n\n\n\n\nNov 17, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nSequencing - Y2H colony PCRs from 20081112\n\n\n\n\n\n\n\n2008\n\n\nMyostatin Interacting Proteins\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nMass Spec - Band #1 from 20081106\n\n\n\n\n\n\n\n2008\n\n\nMyostatin Interacting Proteins\n\n\n\n\n\n\n\n\n\n\n\nNov 14, 2008\n\n\nSam White\n\n\n\n\n\n\n \n\n\n\n\nWestern Blot - Purified (His column) decorin, FST, LAP & telethonin\n\n\n\n\n\n\n\n2008\n\n\nMyostatin Interacting Proteins\n\n\n\n\n\n\n\n\n\n\n\nNov 13, 2008\n\n\nSam White\n\n\n\n\n\n\nNo matching items"
},
{
"objectID": "posts/2024/2024-01-02-Daily-Bits---January-2024/index.html#section-3",
@@ -11023,5 +11023,12 @@
"title": "Daily Bits - January 2024",
"section": "20240102",
"text": "20240102"
+ },
+ {
+ "objectID": "posts/2024/2024-01-03-Data-Exploration---CEABIGR-Spurious-Transcription-Calculations-and-Plotting/index.html",
+ "href": "posts/2024/2024-01-03-Data-Exploration---CEABIGR-Spurious-Transcription-Calculations-and-Plotting/index.html",
+ "title": "Data Exploration - CEABIGR Spurious Transcription Calculations and Plotting",
+ "section": "",
+ "text": "Intro\nAs part of the CEABIGR project (GitHub repo) Steven performed some inital data wrangling to test out the basic calculations to determine the natural log of the fold change in exon expression, relative to Exon 1, for each gene in a single sample. The decision to perform the calculation in this manner was based on (Li et al. 2018). I took the next steps to perform this across all samples, as well as generate some comparison plots. After this, will explore how spurious transcription relates to methylation levels across genes.\nSee 65-exon-coverage.qmd for code.\n\n\n\n\n\n\nNote\n\n\n\nCode and plots link to commit 8bc64a3.\n\n\n\n\nPlots\nAll plots are line plots of the mean natural log fold-change in exon expression (Exons 2-6), relative to Exon 1. Black bars represent standard error.\n\n\n\n\n\n\nNote\n\n\n\nPlots are simply arranged side-by-side. Scales of axes are not intended to match.\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\nReferences\n\nLi, Yong, Yi Jin Liew, Guoxin Cui, Maha J. Cziesielski, Noura Zahran, Craig T. Michell, Christian R. Voolstra, and Manuel Aranda. 2018. “DNA Methylation Regulates Transcriptional Homeostasis of Algal Endosymbiosis in the Coral Model Aiptasia.” Science Advances 4 (8). https://doi.org/10.1126/sciadv.aat2142."
}
]
\ No newline at end of file
diff --git a/docs/sitemap.xml b/docs/sitemap.xml
index 446b6f6a6..4f11c3cdd 100644
--- a/docs/sitemap.xml
+++ b/docs/sitemap.xml
@@ -5986,6 +5986,10 @@
https://robertslab.github.io/sams-notebook/index.html
- 2024-01-10T23:08:43.282Z
+ 2024-01-10T23:18:15.486Z
+
+
+ https://robertslab.github.io/sams-notebook/posts/2024/2024-01-03-Data-Exploration---CEABIGR-Spurious-Transcription-Calculations-and-Plotting/index.html
+ 2024-01-10T23:13:02.654Z