diff --git a/docs/Change_Log.md b/docs/Change_Log.md
index c7ea058..ae279dc 100644
--- a/docs/Change_Log.md
+++ b/docs/Change_Log.md
@@ -1,7 +1,8 @@
# Change Log
-* * X.X.X (July, X, 2024)
+* 0.8.2 (July, 10, 2024)
* BUG FIX: Increased checking to avoid javascript errors in Jupyter Lab
and Notebook >= 7, while maintaining NBClassic capabilities.
+ * Documentation updates and improvements.
* 0.8.1 (May 2, 2023)
* BUG FIX: Now always waits for all data to be transferred to plot before
writing backup to a file.
diff --git a/docs/Installation.md b/docs/Installation.md
index 23dc0c1..162132f 100644
--- a/docs/Installation.md
+++ b/docs/Installation.md
@@ -129,16 +129,16 @@ Python](https://docs.python-guide.org/dev/virtualenvs/).
https://cms.gutow.uwosh.edu/Gutow/useful-chemistry-links/software-tools-and-coding/computer-and-coding-how-tos/installing-jupyter-on-raspberrian)
for a discussion of adding swap space on a Pi.
6. Still within the environment shell test
- this by starting jupyter `jupyter notebook` or `jupyter lab`. Jupyter should
- launch in your browser.
+ this by starting jupyter `jupyter notebook`, `jupyter lab` or `jupyter
+ nbclassic`. Jupyter should launch in your browser.
1. Open a new notebook using the default (Python 3) kernel.
2. In the first cell import all from DAQinstance.py:
`from jupyterpidaq.DAQinstance import *`.
- When run in jupyter notebook this cell should load the DAQmenu at the
- end of the Jupyter notebook menu/icon bar. Currently, no convenience
- menu is available in Jupyter lab. If you do not have an appropriate
- A-to-D board installed you will get a message and the software
- will default to demo mode, substituting a random number
+ In classic notebook this cell should load the DAQmenu at the
+ end of the Jupyter notebook menu/icon bar. In Notebook and Lab DAQ
+ Commands menu should appear on launch of Jupyter. If you do not
+ have an appropriate A-to-D board installed you will get a message and
+ the software will default to demo mode, substituting a random number
generator for the A-to-D.
7. If you wish, you can make this environment available to an alternate Jupyter
install as a special kernel when you are the user.
diff --git a/docs/User_Guide.md b/docs/User_Guide.md
index e59b780..4f6ab4d 100644
--- a/docs/User_Guide.md
+++ b/docs/User_Guide.md
@@ -42,8 +42,9 @@ up, that lead to slightly different steps for starting the software:
[Installation Instructions](https://jupyterphysscilab.github.io/JupyterPiDAQ/jupyterpidaq.html#installation)).
* In this case launch
Jupyter, in whichever directory you want to work, using the
- command: `jupyter nbclassic` (for the old notebook interface) or `jupyter
- lab` (for the newer lab interface).
+ command: `jupyter lab` (for the newer lab interface), `jupyter notebook`
+ (for the modern notebook interface), or `jupyter nbclassic` (for the old
+ notebook interface).
* Open a new notebook and choose the kernel
for `JupyterPiDAQ`. The kernel name will depend upon what was chosen
during installation.
@@ -52,25 +53,29 @@ up, that lead to slightly different steps for starting the software:
* In this case you must navigate to the
directory of the virtual environment using the `cd` command before
starting the software.
- * Then enter the virtual environment with the command `pipenv shell`.
- This assumes you set up `pipenv` as described in the
- [Installation instructions](https://jupyterphysscilab.github.io/JupyterPiDAQ/jupyterpidaq.html#installation).
- * Launch Jupyter using the command: `jupyter nbclassic` (for the old
- notebook interface) or `jupyter lab` (for the newer lab interface).
+ * Then enter the virtual environment with the command `pipenv shell` (you
+ might be using `venv` or something else). This assumes you set up
+ `pipenv` as described in the [Installation instructions
+ ](https://jupyterphysscilab.github.io/JupyterPiDAQ/jupyterpidaq.html#installation).
+ * Launch Jupyter using the
+ command: `jupyter lab` (for the newer lab interface), `jupyter notebook`
+ (for the modern notebook interface), or `jupyter nbclassic` (for the old
+ notebook interface).
* Open a new python notebook.
### Initialize the Data Acquisition Tools
Initialize the data acquisition tools by putting the statement `from
-jupyterpidaq.DAQinstance import *` into the first cell and clicking on the
-'Run' button. The package loads supporting packages (numpy, pandas, plotly,
-etc...) and searches for compatible hardware. On a Raspberry Pi this takes a
-number of seconds. If no compatible A-to-D boards are found, demo mode is used.
-In demo mode the A-to-D board is simulated by a random number generator.
+jupyterpidaq.DAQinstance import *` into the first cell and then running
+(executing) the cell. The package loads supporting packages (numpy, pandas,
+plotly, etc...) and searches for compatible hardware. On a Raspberry Pi this
+takes a number of seconds. If no compatible A-to-D boards are found, demo
+mode is used. In demo mode the A-to-D board is simulated by a random number
+generator.
When setup is done a new menu appears at the end of the menubar (figure 1).
-
+
**Figure 1**: The menu created once the data acquisition software is
initialized. Currently NOT available in Jupyter Lab.
@@ -82,11 +87,10 @@ a new column in a DataFrame, or fitting data.
### Collecting data
**Using the Menu**
-From the "DAQ Commands" menu select "Insert New Run After Selection..." or
-"Append New Run to End...". The first will insert the code to set up
-data collection for a run in the cell below the currently selected cell.
-The second will append this to the end of the notebook. When the cell is
-run it will generate
+From the "DAQ Commands" menu select "Insert New run after selected cell..."
+This will insert the code to set up data collection for a run in the cell
+below the currently selected cell. Insert the name you want for the run
+into the code as specified. When the cell is run it will generate
a GUI that looks like the figure 2, below. Fill in the information to define
what you wish to do (see below figure 2 for more details).
@@ -98,10 +102,10 @@ string you want for this run. Replacing spaces with _ will prevent issues,
especially on Windows. After entering the command, run the cell. This will
generate the GUI shown in figure 2.
- +
+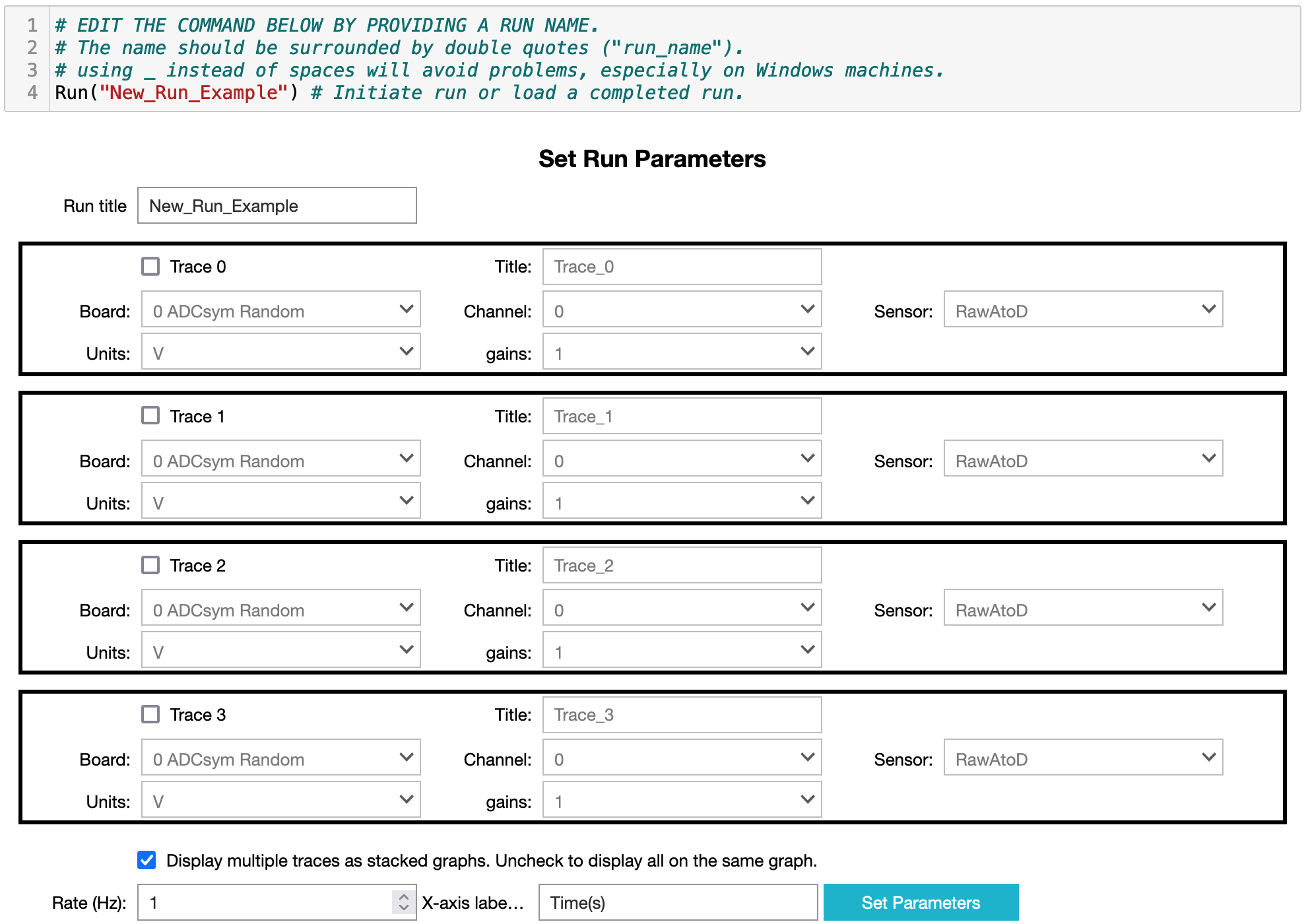 **Figure 2**: Image of the GUI for setting up a data collection run.
-1. Give the run a title/name using the first textbox.
+1. You can give the run a different title/name using the first textbox.
2. You can collect up to four (4) data traces at once. Two different data
traces can display the same analog-to-digital channel, but in different
units (e.g. you could record both the raw voltage and temperature from a
@@ -135,21 +139,20 @@ generate the GUI shown in figure 2.
#### In a table
-Selecting the "Show data in table..." option in the "DAQ Command" menu will
-insert a cell immediately below the currently selected cell displaying a
-widget in which you can select which data set to display. The command
-equivalent is `showDataTable()`.
+Selecting the "Select data to show in a table..." option in the "DAQ
+Command" menu will insert a cell immediately below the currently selected
+cell displaying a widget in which you can select which data set to display.
+The command equivalent is `showDataTable()`.
#### Plotting data
-Selecting the "Insert new plot after selection..." option in the menu will
-insert two cells immediately below the currently selected cell. The first cell
-will be used to generate a GUI to lead you through creation of the code to
-generate the plot. The second cell is where the plot creation code is
-generated. The first tab of this GUI looks like figure 3. The command
-equivalent is `newPlot()`.
+Selecting the "Insert new plot after selected cell" option in the menu will
+insert a cell immediately below the currently selected cell. This will
+create a GUI to lead you through generating code to make a plot of a data
+set. The code is generated immediately below the GUI. The first tab of this GUI
+looks like figure 3. The command equivalent is `newPlot()`.
-
+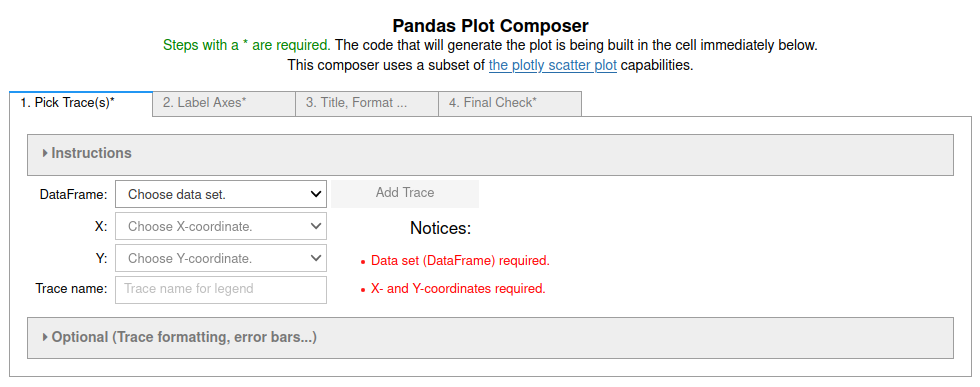
**Figure 3**: Image of the first tab in the four tab (4 step) Pandas Plot
Composer. More information in the [Pandas_GUI
@@ -170,13 +173,13 @@ The GUI destroys itself once you complete step 4.
#### Calculating a new column
-Selecting "Calculate new column..." from the menu will add two cells
-immediately below the selected cell. The first cell will create the GUI to
-lead you through creation of the code to calculate the new column. The second
-cell is where the code is built. The first tab of the GUI looks like
+Selecting "Calculate new column..." from the menu will a cell
+immediately below the selected cell and launch the GUI to
+lead you through creation of the code to calculate the new column. The code
+is created immediately below the GUI. The first tab of the GUI looks like
figure 4. The command equivalent is `newCalculatedColumn()`.
-
+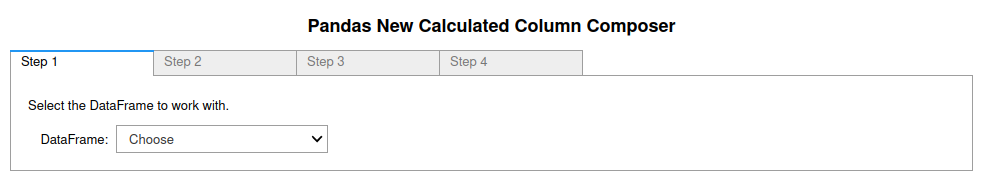
**Figure 4**: Image of the first tab in the four tab (4 step) Pandas New
Calculated Column Composer. More information in the [Pandas_GUI
@@ -190,12 +193,12 @@ The GUI destroys itself once you complete step 4.
#### Fitting data
A GUI for defining simple fits (linear, polynomial, exponential decay, sine
-and Gaussian) can be launched by selecting "Insert new Fit after selection..
-." from the menu. This will create the GUI to lead you through selecting
-and fitting the data. The code is created in the cell immediately below the
+and Gaussian) can be launched by selecting "Insert new fit after selected
+cell" from the menu. This will create the GUI to lead you through selecting
+and fitting the data. The code is created the immediately below the
GUI. The command equivalent is `newFit()`.
-
+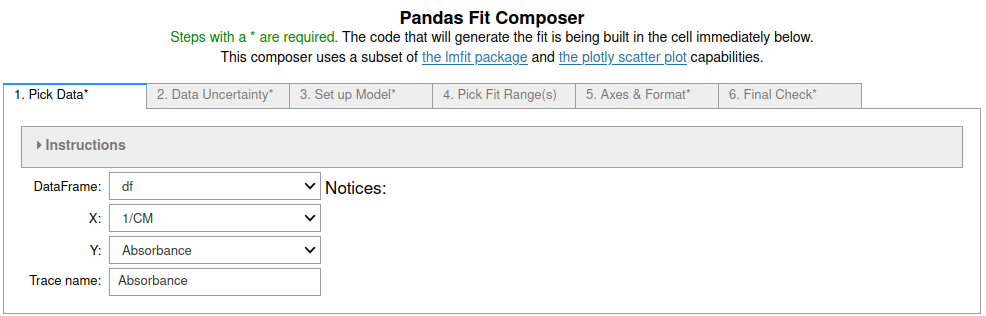
**Figure 5**: Image of the first tab in the Pandas Fit Composer.
More information in the [Pandas_GUI
**Figure 2**: Image of the GUI for setting up a data collection run.
-1. Give the run a title/name using the first textbox.
+1. You can give the run a different title/name using the first textbox.
2. You can collect up to four (4) data traces at once. Two different data
traces can display the same analog-to-digital channel, but in different
units (e.g. you could record both the raw voltage and temperature from a
@@ -135,21 +139,20 @@ generate the GUI shown in figure 2.
#### In a table
-Selecting the "Show data in table..." option in the "DAQ Command" menu will
-insert a cell immediately below the currently selected cell displaying a
-widget in which you can select which data set to display. The command
-equivalent is `showDataTable()`.
+Selecting the "Select data to show in a table..." option in the "DAQ
+Command" menu will insert a cell immediately below the currently selected
+cell displaying a widget in which you can select which data set to display.
+The command equivalent is `showDataTable()`.
#### Plotting data
-Selecting the "Insert new plot after selection..." option in the menu will
-insert two cells immediately below the currently selected cell. The first cell
-will be used to generate a GUI to lead you through creation of the code to
-generate the plot. The second cell is where the plot creation code is
-generated. The first tab of this GUI looks like figure 3. The command
-equivalent is `newPlot()`.
+Selecting the "Insert new plot after selected cell" option in the menu will
+insert a cell immediately below the currently selected cell. This will
+create a GUI to lead you through generating code to make a plot of a data
+set. The code is generated immediately below the GUI. The first tab of this GUI
+looks like figure 3. The command equivalent is `newPlot()`.
-
+
**Figure 3**: Image of the first tab in the four tab (4 step) Pandas Plot
Composer. More information in the [Pandas_GUI
@@ -170,13 +173,13 @@ The GUI destroys itself once you complete step 4.
#### Calculating a new column
-Selecting "Calculate new column..." from the menu will add two cells
-immediately below the selected cell. The first cell will create the GUI to
-lead you through creation of the code to calculate the new column. The second
-cell is where the code is built. The first tab of the GUI looks like
+Selecting "Calculate new column..." from the menu will a cell
+immediately below the selected cell and launch the GUI to
+lead you through creation of the code to calculate the new column. The code
+is created immediately below the GUI. The first tab of the GUI looks like
figure 4. The command equivalent is `newCalculatedColumn()`.
-
+
**Figure 4**: Image of the first tab in the four tab (4 step) Pandas New
Calculated Column Composer. More information in the [Pandas_GUI
@@ -190,12 +193,12 @@ The GUI destroys itself once you complete step 4.
#### Fitting data
A GUI for defining simple fits (linear, polynomial, exponential decay, sine
-and Gaussian) can be launched by selecting "Insert new Fit after selection..
-." from the menu. This will create the GUI to lead you through selecting
-and fitting the data. The code is created in the cell immediately below the
+and Gaussian) can be launched by selecting "Insert new fit after selected
+cell" from the menu. This will create the GUI to lead you through selecting
+and fitting the data. The code is created the immediately below the
GUI. The command equivalent is `newFit()`.
-
+
**Figure 5**: Image of the first tab in the Pandas Fit Composer.
More information in the [Pandas_GUI
 +
+ **Figure 2**: Image of the GUI for setting up a data collection run.
-1. Give the run a title/name using the first textbox.
+1. You can give the run a different title/name using the first textbox.
2. You can collect up to four (4) data traces at once. Two different data
traces can display the same analog-to-digital channel, but in different
units (e.g. you could record both the raw voltage and temperature from a
@@ -135,21 +139,20 @@ generate the GUI shown in figure 2.
#### In a table
-Selecting the "Show data in table..." option in the "DAQ Command" menu will
-insert a cell immediately below the currently selected cell displaying a
-widget in which you can select which data set to display. The command
-equivalent is `showDataTable()`.
+Selecting the "Select data to show in a table..." option in the "DAQ
+Command" menu will insert a cell immediately below the currently selected
+cell displaying a widget in which you can select which data set to display.
+The command equivalent is `showDataTable()`.
#### Plotting data
-Selecting the "Insert new plot after selection..." option in the menu will
-insert two cells immediately below the currently selected cell. The first cell
-will be used to generate a GUI to lead you through creation of the code to
-generate the plot. The second cell is where the plot creation code is
-generated. The first tab of this GUI looks like figure 3. The command
-equivalent is `newPlot()`.
+Selecting the "Insert new plot after selected cell" option in the menu will
+insert a cell immediately below the currently selected cell. This will
+create a GUI to lead you through generating code to make a plot of a data
+set. The code is generated immediately below the GUI. The first tab of this GUI
+looks like figure 3. The command equivalent is `newPlot()`.
-
+
**Figure 3**: Image of the first tab in the four tab (4 step) Pandas Plot
Composer. More information in the [Pandas_GUI
@@ -170,13 +173,13 @@ The GUI destroys itself once you complete step 4.
#### Calculating a new column
-Selecting "Calculate new column..." from the menu will add two cells
-immediately below the selected cell. The first cell will create the GUI to
-lead you through creation of the code to calculate the new column. The second
-cell is where the code is built. The first tab of the GUI looks like
+Selecting "Calculate new column..." from the menu will a cell
+immediately below the selected cell and launch the GUI to
+lead you through creation of the code to calculate the new column. The code
+is created immediately below the GUI. The first tab of the GUI looks like
figure 4. The command equivalent is `newCalculatedColumn()`.
-
+
**Figure 4**: Image of the first tab in the four tab (4 step) Pandas New
Calculated Column Composer. More information in the [Pandas_GUI
@@ -190,12 +193,12 @@ The GUI destroys itself once you complete step 4.
#### Fitting data
A GUI for defining simple fits (linear, polynomial, exponential decay, sine
-and Gaussian) can be launched by selecting "Insert new Fit after selection..
-." from the menu. This will create the GUI to lead you through selecting
-and fitting the data. The code is created in the cell immediately below the
+and Gaussian) can be launched by selecting "Insert new fit after selected
+cell" from the menu. This will create the GUI to lead you through selecting
+and fitting the data. The code is created the immediately below the
GUI. The command equivalent is `newFit()`.
-
+
**Figure 5**: Image of the first tab in the Pandas Fit Composer.
More information in the [Pandas_GUI
**Figure 2**: Image of the GUI for setting up a data collection run.
-1. Give the run a title/name using the first textbox.
+1. You can give the run a different title/name using the first textbox.
2. You can collect up to four (4) data traces at once. Two different data
traces can display the same analog-to-digital channel, but in different
units (e.g. you could record both the raw voltage and temperature from a
@@ -135,21 +139,20 @@ generate the GUI shown in figure 2.
#### In a table
-Selecting the "Show data in table..." option in the "DAQ Command" menu will
-insert a cell immediately below the currently selected cell displaying a
-widget in which you can select which data set to display. The command
-equivalent is `showDataTable()`.
+Selecting the "Select data to show in a table..." option in the "DAQ
+Command" menu will insert a cell immediately below the currently selected
+cell displaying a widget in which you can select which data set to display.
+The command equivalent is `showDataTable()`.
#### Plotting data
-Selecting the "Insert new plot after selection..." option in the menu will
-insert two cells immediately below the currently selected cell. The first cell
-will be used to generate a GUI to lead you through creation of the code to
-generate the plot. The second cell is where the plot creation code is
-generated. The first tab of this GUI looks like figure 3. The command
-equivalent is `newPlot()`.
+Selecting the "Insert new plot after selected cell" option in the menu will
+insert a cell immediately below the currently selected cell. This will
+create a GUI to lead you through generating code to make a plot of a data
+set. The code is generated immediately below the GUI. The first tab of this GUI
+looks like figure 3. The command equivalent is `newPlot()`.
-
+
**Figure 3**: Image of the first tab in the four tab (4 step) Pandas Plot
Composer. More information in the [Pandas_GUI
@@ -170,13 +173,13 @@ The GUI destroys itself once you complete step 4.
#### Calculating a new column
-Selecting "Calculate new column..." from the menu will add two cells
-immediately below the selected cell. The first cell will create the GUI to
-lead you through creation of the code to calculate the new column. The second
-cell is where the code is built. The first tab of the GUI looks like
+Selecting "Calculate new column..." from the menu will a cell
+immediately below the selected cell and launch the GUI to
+lead you through creation of the code to calculate the new column. The code
+is created immediately below the GUI. The first tab of the GUI looks like
figure 4. The command equivalent is `newCalculatedColumn()`.
-
+
**Figure 4**: Image of the first tab in the four tab (4 step) Pandas New
Calculated Column Composer. More information in the [Pandas_GUI
@@ -190,12 +193,12 @@ The GUI destroys itself once you complete step 4.
#### Fitting data
A GUI for defining simple fits (linear, polynomial, exponential decay, sine
-and Gaussian) can be launched by selecting "Insert new Fit after selection..
-." from the menu. This will create the GUI to lead you through selecting
-and fitting the data. The code is created in the cell immediately below the
+and Gaussian) can be launched by selecting "Insert new fit after selected
+cell" from the menu. This will create the GUI to lead you through selecting
+and fitting the data. The code is created the immediately below the
GUI. The command equivalent is `newFit()`.
-
+
**Figure 5**: Image of the first tab in the Pandas Fit Composer.
More information in the [Pandas_GUI